Pseudoxanthomonas winnipegensis: Difference between revisions
| (30 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | {{Uncurated}} | ||
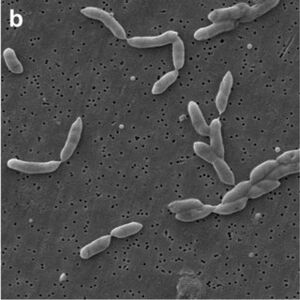
[[Image: | [[Image:Pseudoxanthomonaswinnipegensis.jpg|thumb|300px|right|Legend. Image credit: International Journal of Systematic and Evolutionary Microbiology.]] | ||
| Line 19: | Line 19: | ||
| height="10" bgcolor="#FFDF95" | | | height="10" bgcolor="#FFDF95" | | ||
'''NCBI: [https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id= | '''NCBI: [https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=2480810&lvl=3&lin=f&keep=1&srchmode=1&unlock]''' | ||
|} | |} | ||
''Pseudoxanthomonas winnipegensis'' | |||
''Pseudoxanthomonas'' | |||
==Description and Significance== | ==Description and Significance== | ||
Pseudoxanthomonas winnipegensis is a gram negative bacteria. This species obtains the bacillus shape and has a yellowish pigment. Pseudoxanthomonas winnipegensis was derived water, plants | <i>Pseudoxanthomonas winnipegensis</i> is a gram negative bacteria. This species obtains the bacillus shape and has a yellowish pigment. <i>Pseudoxanthomonas winnipegensis</i> was derived from water, plants and contaminated soils. Recent studies recovered the bacteria from human clinical materials and was recently recovered from patients with cystic fibrosis. Until recently, Pseudoxanthomonas species were only identifiable at the genus level. However, new gene sequencing improvements have allowed for the identification of the species within the genus level. | ||
==Genome Structure== | ==Genome Structure== | ||
Specific strains of this bacteria were studied by whole genome sequencing which used average nucleotide identity, DNA-DNA hybridization, amino acid identity, core genome and nucleotide analyses, biochemical and cellular fatty acid analysis and antimicrobial susceptibility testing. Pseudoxanthomonas | Specific strains of this bacteria were studied by whole genome sequencing which used average nucleotide identity, DNA-DNA hybridization, amino acid identity, core genome and nucleotide analyses, biochemical and cellular fatty acid analysis and antimicrobial susceptibility testing. The structure of <i>Pseudoxanthomonas winnipegensis</i> was investigated using scanning microscopes and transmission electron microscopes. The bacteria was found to be a gram-negative bacilli. Genome sequencing was analyzed using 16S rRNA. Findings showed the bacteria had a average genome size ranging from 4.36 to 4.73 Mb. The genome also has an average G+C content of 69.12 mol%. Additionally, genome sequencing found <i>Pseudoxanthomonas winnipegensis</i> has a circular chromosomal structure with 4,505,034 bases, and a CDS of 4077. | ||
==Cell Structure, Metabolism and Life Cycle== | ==Cell Structure, Metabolism and Life Cycle== | ||
Pseudoxanthomonas winnipegensis is gram-negative bacilli with a yellowish pigment. Additionally, research has found they are aerobes, oxidative and non-motile. The bacteria is susceptible to most antibiotics. In the laboratory, <i>Pseudoxanthomonas winnipegensis</i> isolates had 0.5 mm −1 mm, convex, yellowish-beige colonies when grown on agar. The colonies are medium-length, slightly curved and asporogenous. It is able to oxidize/assimilate glucose, lactose, sucrose, maltose and fructose. | |||
==Ecology and Pathogenesis== | ==Ecology and Pathogenesis== | ||
<i>Pseudoxanthomonas winnipegensis</i> obtained its name from Winnipeg Manitoba Canada, where the species strains were characterized. <i>Pseudoxanthomonas winnipegensis</i> grows at an optimal temperature of 35–37 °C in 5 % CO2. Cultures were found to have limited growth at 25 °C or 42 °C. The bacteria does grow aerobically, however does not grow anaerobically. Additionally, <i>Pseudoxanthomonas winnipegensis</i> is catalase positive and is able to oxidize glucose, lactose, sucrose, maltose, and fructose. New research found strains of this bacteria recovered from ear and throat samples from patients with cystic fibrosis, specifically from ear infections, dialysate and a sinus specimen. There is a lack of information on P. winnipegensis at the time due to the recency of its discovery, however similar Pseudoxanthomonas species found in rhizosphere soil possessed a broad spectrum of antimicrobial activity against pathogenic microorganisms. | |||
==References== | ==References== | ||
[ | [Bernard, K. A., Vachon, A., Pacheco, A. L., Burdz, T., Wiebe, D., Beniac, D. R., Hiebert, S. L., Booth, T., Doyle, D. A., Lawson, P., & Bernier, A. M. (2020). Pseudoxanthomonas winnipegensis sp. nov., derived from human clinical materials and recovered from cystic fibrosis and other patient types in Canada, and emendation of Pseudoxanthomonas spadix Young et al. 2007. International journal of systematic and evolutionary microbiology, 70(12), 6313–6322. https://doi.org/10.1099/ijsem.0.004533.] | ||
[Nayaka, S. 2019. Isolation, characterization, and functional groups analysis of Pseudoxanthomonas indica RSA-23 from rhizosphere soil. Journal of Applied Pharmaceutical Science Vol. 9(11), pp 101-106. DOI: 10.7324/JAPS.2019.91113] | |||
==Author== | ==Author== | ||
Latest revision as of 22:36, 13 December 2022
Classification
Domain;Bacteria Phylum;Pseudomonadota Class;Gammaproteobacteria Order;Xanthomonadales Family; Xanthomonadaceae
Species
|
NCBI: [1] |
Pseudoxanthomonas winnipegensis
Description and Significance
Pseudoxanthomonas winnipegensis is a gram negative bacteria. This species obtains the bacillus shape and has a yellowish pigment. Pseudoxanthomonas winnipegensis was derived from water, plants and contaminated soils. Recent studies recovered the bacteria from human clinical materials and was recently recovered from patients with cystic fibrosis. Until recently, Pseudoxanthomonas species were only identifiable at the genus level. However, new gene sequencing improvements have allowed for the identification of the species within the genus level.
Genome Structure
Specific strains of this bacteria were studied by whole genome sequencing which used average nucleotide identity, DNA-DNA hybridization, amino acid identity, core genome and nucleotide analyses, biochemical and cellular fatty acid analysis and antimicrobial susceptibility testing. The structure of Pseudoxanthomonas winnipegensis was investigated using scanning microscopes and transmission electron microscopes. The bacteria was found to be a gram-negative bacilli. Genome sequencing was analyzed using 16S rRNA. Findings showed the bacteria had a average genome size ranging from 4.36 to 4.73 Mb. The genome also has an average G+C content of 69.12 mol%. Additionally, genome sequencing found Pseudoxanthomonas winnipegensis has a circular chromosomal structure with 4,505,034 bases, and a CDS of 4077.
Cell Structure, Metabolism and Life Cycle
Pseudoxanthomonas winnipegensis is gram-negative bacilli with a yellowish pigment. Additionally, research has found they are aerobes, oxidative and non-motile. The bacteria is susceptible to most antibiotics. In the laboratory, Pseudoxanthomonas winnipegensis isolates had 0.5 mm −1 mm, convex, yellowish-beige colonies when grown on agar. The colonies are medium-length, slightly curved and asporogenous. It is able to oxidize/assimilate glucose, lactose, sucrose, maltose and fructose.
Ecology and Pathogenesis
Pseudoxanthomonas winnipegensis obtained its name from Winnipeg Manitoba Canada, where the species strains were characterized. Pseudoxanthomonas winnipegensis grows at an optimal temperature of 35–37 °C in 5 % CO2. Cultures were found to have limited growth at 25 °C or 42 °C. The bacteria does grow aerobically, however does not grow anaerobically. Additionally, Pseudoxanthomonas winnipegensis is catalase positive and is able to oxidize glucose, lactose, sucrose, maltose, and fructose. New research found strains of this bacteria recovered from ear and throat samples from patients with cystic fibrosis, specifically from ear infections, dialysate and a sinus specimen. There is a lack of information on P. winnipegensis at the time due to the recency of its discovery, however similar Pseudoxanthomonas species found in rhizosphere soil possessed a broad spectrum of antimicrobial activity against pathogenic microorganisms.
References
[Bernard, K. A., Vachon, A., Pacheco, A. L., Burdz, T., Wiebe, D., Beniac, D. R., Hiebert, S. L., Booth, T., Doyle, D. A., Lawson, P., & Bernier, A. M. (2020). Pseudoxanthomonas winnipegensis sp. nov., derived from human clinical materials and recovered from cystic fibrosis and other patient types in Canada, and emendation of Pseudoxanthomonas spadix Young et al. 2007. International journal of systematic and evolutionary microbiology, 70(12), 6313–6322. https://doi.org/10.1099/ijsem.0.004533.]
[Nayaka, S. 2019. Isolation, characterization, and functional groups analysis of Pseudoxanthomonas indica RSA-23 from rhizosphere soil. Journal of Applied Pharmaceutical Science Vol. 9(11), pp 101-106. DOI: 10.7324/JAPS.2019.91113]
Author
Page authored by Bella Manfredi, student of Prof. Bradley Tolar at UNC Wilmington.