Alcoholism and its Effects on Gut Microbiome: Difference between revisions
| Line 9: | Line 9: | ||
==How to Analyze the Intestinal Microbial Population?== | ==How to Analyze the Intestinal Microbial Population?== | ||
consider how researchers can evaluate the number of microorganisms in the microbial populations and monitor changes in its size and composition. This is not exactly an easy thing to do, as directly measuring microbial communities such as those within the GIT comes along with certain challenges and ambiguities. Firstly, microorganisms in the gut environment maintain unbelievable genetic diversity but store this diversity in a very limited array of cellular morphologies.<ref name=Woese>[https://doi.org/10.1128/mr.51.2.221-271.1987 Woese C. R. (1987). Bacterial evolution. Microbiological reviews, 51(2), 221–271.]</ref>Additionally, microorganisms have unessential functions that complicate the analysis, share some of the functional capabilities with closely related microorganisms, and most importantly, are extraordinarily difficult to isolate under laboratory conditions.<ref name=Woese></ref>Consideration of all these factors leads to strong preference towards the use of molecular tools - tools that look for and examine DNA and RNA related to specific microorganisms. This allows for careful analysis of these complex communities. Most commonly, the use of one of the two most popular techniques is utilized. These are polymerase chain reaction (PCR) and shotgun sequencing. | consider how researchers can evaluate the number of microorganisms in the microbial populations and monitor changes in its size and composition. This is not exactly an easy thing to do, as directly measuring microbial communities such as those within the GIT comes along with certain challenges and ambiguities. Firstly, microorganisms in the gut environment maintain unbelievable genetic diversity but store this diversity in a very limited array of cellular morphologies.<ref name=Woese>[https://doi.org/10.1128/mr.51.2.221-271.1987 Woese C. R. (1987). Bacterial evolution. Microbiological reviews, 51(2), 221–271.]</ref>Additionally, microorganisms have unessential functions that complicate the analysis, share some of the functional capabilities with closely related microorganisms, and most importantly, are extraordinarily difficult to isolate under laboratory conditions.<ref name=Woese></ref>Consideration of all these factors leads to strong preference towards the use of molecular tools - tools that look for and examine DNA and RNA related to specific microorganisms. This allows for careful analysis of these complex communities. Most commonly, the use of one of the two most popular techniques is utilized. These are polymerase chain reaction (PCR) and shotgun sequencing. | ||
==PCR and Total Bacterial Quantification== | |||
==Alcohol as a Culprit of Dysbiosis in the Intestinal Microbiota== | ==Alcohol as a Culprit of Dysbiosis in the Intestinal Microbiota== | ||
Revision as of 22:35, 11 April 2023
Introduction
By Nikola Kovacova
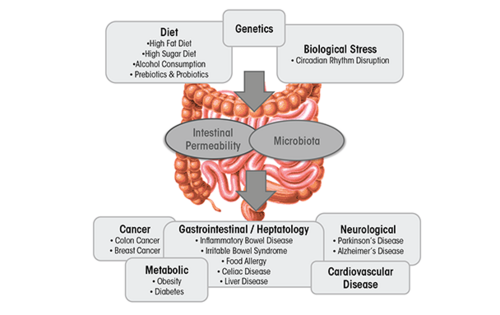
The gut microbiota is classified as a collection of all microbial organisms within the gastrointestinal tract (GIT).[1] With trillions of microorganisms inhabiting the microbiota, its collective genome is believed to encode 100 times more genes than the human genome.[2]This makes it not only a complex, but also an essential player involved in the health status of the GIT, as it impacts digestion, inflammation, and immunity.[3]The microbiota–host relationship is mutualistic - the microbiota contributes to the extraction of energy from various sources, boosts synthesis of vitamins and amino acids, and assists with the formation of barriers against pathogens.[4]This relationship, however, is highly sensitive towards imbalances in microbial composition that can often lead to a decrease in overall well-being of the host.[5]This imbalance, referred to as dysbiosis, can be easily caused by environmental factors incorporated into the lives of countless people in contemporary Western societies, especially their diet. Diets high in fat and sugar have serious negative consequences on the microbiota composition and diversity, contributing to the development of obesity and liver injury, as well as multiple diseases such as IBD, IBS, celiac disease, type 1 and type 2 diabetes, food allergies, and cardiovascular disease.[5]
This page focuses on one specific dietary disruptor of the gut microbiota, namely. Alcohol. Alcoholism is a worldwide problem with implications far beyond the healthcare sector, as drastic social, financial, and interpersonal consequences in the life of the person stricken with alcoholism often occur. In 2018, The World Health Organization has revealed that excessive alcohol consumption is responsible for 3 million deaths yearly and over astonishing 280 million people globally have a diagnosed alcohol use disorder. It has been well-established that the toxicity of the alcohol itself is associated with formation of various health complications. These might be tissue injury and organ dysfunction, higher risk of developing cancer, or abnormal function of the immune system that increases the risk of acute and chronic infections.[6]This review is interested in addressing alcohol’s specific impact on the microbiota in the gastrointestinal tract (GIT), the compositional and functional changes it induces, and corresponding health-related issues.
How to Analyze the Intestinal Microbial Population?
consider how researchers can evaluate the number of microorganisms in the microbial populations and monitor changes in its size and composition. This is not exactly an easy thing to do, as directly measuring microbial communities such as those within the GIT comes along with certain challenges and ambiguities. Firstly, microorganisms in the gut environment maintain unbelievable genetic diversity but store this diversity in a very limited array of cellular morphologies.[7]Additionally, microorganisms have unessential functions that complicate the analysis, share some of the functional capabilities with closely related microorganisms, and most importantly, are extraordinarily difficult to isolate under laboratory conditions.[7]Consideration of all these factors leads to strong preference towards the use of molecular tools - tools that look for and examine DNA and RNA related to specific microorganisms. This allows for careful analysis of these complex communities. Most commonly, the use of one of the two most popular techniques is utilized. These are polymerase chain reaction (PCR) and shotgun sequencing.
PCR and Total Bacterial Quantification
Alcohol as a Culprit of Dysbiosis in the Intestinal Microbiota
Include some current research, with at least one figure showing data.
Alcohol-Induced Diseases of the GIT
Possible Treatments and Therapeutic Interventions
Conclusion
References
- ↑ Savage, D.C. "Microbial ecology of the gastrointestinal tract." Annual review of microbiology, 31, 107–133.
- ↑ Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., Nielsen, T., Pons, N., Levenez, F., Yamada, T., Mende, D. R., Li, J., Xu, J., Li, S., Li, D., Cao, J., Wang, B., Liang, H., Zheng, H., Xie, Y., … Wang, J. (2010). "A human gut microbial gene catalogue established by metagenomic sequencing." Nature, 464(7285), 59–65.
- ↑ Leclercq, S., de Timary, P., Delzenne, N. M., & Stärkel, P. (2017). "The link between inflammation, bugs, the intestine and the brain in alcohol dependence." Translational psychiatry, 7(2), e1048.
- ↑ Tappenden, K. A., & Deutsch, A. S. (2007). "The physiological relevance of the intestinal microbiota--contributions to human health." Journal of the American College of Nutrition, 26(6), 679S–83S.
- ↑ 5.0 5.1 Kim, B. S., Jeon, Y. S., & Chun, J. (2013). "Current status and future promise of the human microbiome." Pediatric gastroenterology, hepatology & nutrition, 16(2), 71–79.
- ↑ Szabo, G., & Mandrekar, P. (2009). A recent perspective on alcohol, immunity, and host defense. Alcoholism, clinical and experimental research, 33(2), 220–232.
- ↑ 7.0 7.1 Woese C. R. (1987). Bacterial evolution. Microbiological reviews, 51(2), 221–271.
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2023, Kenyon College
