Alcoholism and its Effects on Gut Microbiome: Difference between revisions
| (44 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
<br>By Nikola Kovacova <br> | <br>By Nikola Kovacova <br> | ||
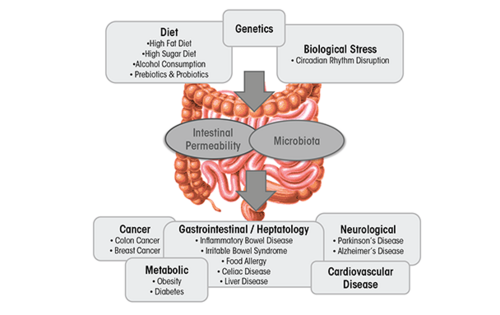
The gut microbiota is classified as a collection of all microbial organisms within the gastrointestinal tract (GIT).<ref name=Savage> [https://doi.org/10.1146/annurev.mi.31.100177.0005430 Savage, D.C. "Microbial ecology of the gastrointestinal tract." Annual review of microbiology, 31, 107–133.]</ref> With trillions of microorganisms inhabiting the microbiota, its collective genome is believed to encode 100 times more genes than the human genome.<ref name=Qin>[https://doi.org/10.1038/nature08821 Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., Nielsen, T., Pons, N., Levenez, F., Yamada, T., Mende, D. R., Li, J., Xu, J., Li, S., Li, D., Cao, J., Wang, B., Liang, H., Zheng, H., Xie, Y., … Wang, J. (2010). "A human gut microbial gene catalogue established by metagenomic sequencing." Nature, 464(7285), 59–65.]</ref> This makes it not only a complex, but also an essential player involved in the health status of the GIT, as it impacts digestion, inflammation, and immunity.<ref name=Leclercq>[https://doi.org/10.1038/tp.2017.15 Leclercq, S., de Timary, P., Delzenne, N. M., & Stärkel, P. (2017). "The link between inflammation, bugs, the intestine and the brain in alcohol dependence." Translational psychiatry, 7(2), e1048.]</ref> The microbiota–host relationship is mutualistic - the microbiota contributes to the extraction of energy from various sources, boosts synthesis of vitamins and amino acids, and assists with the formation of barriers against pathogens.<ref name=Tappenden>[https://doi.org/10.1080/07315724.2007.10719647 Tappenden, K. A., & Deutsch, A. S. (2007). "The physiological relevance of the intestinal microbiota--contributions to human health." Journal of the American College of Nutrition, 26(6), 679S–83S.]</ref> This relationship, however, is highly sensitive towards imbalances in microbial composition that can often lead to a decrease in overall well-being of the host.<ref name=Kim>[https://doi.org/10.5223/pghn.2013.16.2.71 Kim, B. S., Jeon, Y. S., & Chun, J. (2013). "Current status and future promise of the human microbiome." Pediatric gastroenterology, hepatology & nutrition, 16(2), 71–79.]</ref> This imbalance, referred to as dysbiosis, can be easily caused by environmental factors incorporated into the lives of countless people in contemporary Western societies, especially | The gut microbiota is classified as a collection of all microbial organisms within the gastrointestinal tract (GIT).<ref name=Savage> [https://doi.org/10.1146/annurev.mi.31.100177.0005430 Savage, D.C. "Microbial ecology of the gastrointestinal tract." Annual review of microbiology, 31, 107–133.]</ref> With trillions of microorganisms inhabiting the microbiota, its collective genome is believed to encode 100 times more genes than the human genome.<ref name=Qin>[https://doi.org/10.1038/nature08821 Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., Nielsen, T., Pons, N., Levenez, F., Yamada, T., Mende, D. R., Li, J., Xu, J., Li, S., Li, D., Cao, J., Wang, B., Liang, H., Zheng, H., Xie, Y., … Wang, J. (2010). "A human gut microbial gene catalogue established by metagenomic sequencing." Nature, 464(7285), 59–65.]</ref> This makes it not only a complex, but also an essential player involved in the health status of the GIT, as it impacts digestion, inflammation, and immunity.<ref name=Leclercq>[https://doi.org/10.1038/tp.2017.15 Leclercq, S., de Timary, P., Delzenne, N. M., & Stärkel, P. (2017). "The link between inflammation, bugs, the intestine and the brain in alcohol dependence." Translational psychiatry, 7(2), e1048.]</ref> The microbiota–host relationship is mutualistic - the microbiota contributes to the extraction of energy from various sources, boosts synthesis of vitamins and amino acids, and assists with the formation of barriers against pathogens.<ref name=Tappenden>[https://doi.org/10.1080/07315724.2007.10719647 Tappenden, K. A., & Deutsch, A. S. (2007). "The physiological relevance of the intestinal microbiota--contributions to human health." Journal of the American College of Nutrition, 26(6), 679S–83S.]</ref> This relationship, however, is highly sensitive towards imbalances in microbial composition that can often lead to a decrease in overall well-being of the host.<ref name=Kim>[https://doi.org/10.5223/pghn.2013.16.2.71 Kim, B. S., Jeon, Y. S., & Chun, J. (2013). "Current status and future promise of the human microbiome." Pediatric gastroenterology, hepatology & nutrition, 16(2), 71–79.]</ref> This imbalance, referred to as dysbiosis, can be easily caused by environmental factors incorporated into the lives of countless people in the contemporary Western societies, especially the poor dietary choices. Diets high in fat and sugar have serious negative consequences on the microbiota composition and diversity, contributing to the development of obesity and liver injury, as well as multiple diseases such as IBD, IBS, celiac disease, type 1 and type 2 diabetes, food allergies, and cardiovascular disease.<ref name=Kim></ref> | ||
This page focuses on one specific dietary disruptor of the gut microbiota | This page focuses on one specific dietary disruptor of the gut microbiota. Alcohol. Alcoholism is a worldwide problem with implications far beyond the healthcare sector. Drastic social, financial, and interpersonal consequences in the life of the person stricken with alcoholism often occur. The World Health Organization [https://www.who.int/news-room/fact-sheets/detail/alcohol] has revealed that excessive alcohol consumption is responsible for 3 million deaths yearly and over astonishing 280 million people globally have a diagnosed alcohol use disorder. It has been well-established that the toxicity of the alcohol itself is associated with the formation of various health complications. These might be tissue injury and organ dysfunction, higher risk of developing cancer, or abnormal function of the immune system that increases the risk of acute and chronic infections.<ref name=Szabo>[https://doi.org/10.1111/j.1530-0277.2008.00842.x Szabo, G., & Mandrekar, P. (2009). A recent perspective on alcohol, immunity, and host defense. Alcoholism, clinical and experimental research, 33(2), 220–232.]</ref> This review is interested in addressing alcohol’s specific impact on the microbiota in the gastrointestinal tract (GIT), the compositional and functional changes it induces, and corresponding health-related issues. | ||
==How to Analyze the Intestinal Microbial Population?== | ==How to Analyze the Intestinal Microbial Population?== | ||
[[Image:Snip.PNG|thumb|400px|left|Prevalent scientific methods of gut microbiome composition analysis. Figure adapted from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6154731/ ]] | [[Image:Snip.PNG|thumb|400px|left|Prevalent scientific methods of gut microbiome composition analysis. Figure adapted from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6154731/ ]] | ||
Before we dive into the specific alcohol-induced changes in the microbiota, we must first consider how researchers can evaluate the number of microorganisms in the microbial populations and monitor changes in its size and composition. This is not exactly an easy thing to do, as directly measuring microbial communities such as those within the GIT comes along with certain challenges and ambiguities. Firstly, microorganisms in the gut environment maintain unbelievable genetic diversity but store this diversity in a very limited array of cellular morphologies.<ref name=Woese>[https://doi.org/10.1128/mr.51.2.221-271.1987 Woese C. R. (1987). Bacterial evolution. Microbiological reviews, 51(2), 221–271.]</ref> Additionally, microorganisms have unessential functions that complicate the analysis, share some of the functional capabilities with closely related microorganisms, and most importantly | Before we dive into the specific alcohol-induced changes in the microbiota, we must first consider how researchers can evaluate the number of microorganisms in the microbial populations and monitor changes in its size and composition. This is not exactly an easy thing to do, as directly measuring microbial communities such as those within the GIT comes along with certain challenges and ambiguities. Firstly, microorganisms in the gut environment maintain unbelievable genetic diversity but store this diversity in a very limited array of cellular morphologies.<ref name=Woese>[https://doi.org/10.1128/mr.51.2.221-271.1987 Woese C. R. (1987). Bacterial evolution. Microbiological reviews, 51(2), 221–271.]</ref> Additionally, microorganisms have unessential functions that complicate the analysis, share some of the functional capabilities with closely related microorganisms, and most importantly - are extraordinarily difficult to isolate under laboratory conditions.<ref name=Woese></ref> Consideration of all these factors leads to strong preference towards the use of molecular tools - tools that look for and examine DNA and RNA. This allows for careful analysis of these complex microbial communities. Most commonly, the use of one of the two most popular techniques is utilized. These are polymerase chain reaction (PCR) and shotgun sequencing. | ||
===PCR and Total Bacterial Quantification=== | ===PCR and Total Bacterial Quantification=== | ||
Technique of PCR | Technique of PCR consists of walking on a fine line between finding an appropriate gene target, a section that would be frequent enough among microorganisms so that it could be used for searching, but different enough so that it could individuate among microorganisms. Traditionally, utilization of ribosomal RNA (rRNA) genes has been preferred for this molecular analysis. This analysis consists of using PCR-based techniques coupled with indirect fingerprinting or direct sequencing, such as next-generation sequencing (NGS).<ref name=Langille>[https://doi.org/10.1038/nbt.2676 Langille, M., Zaneveld, J., Caporaso, J. et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31, 814–821 (2013).]</ref> This is due to rRNA genes containing very specific DNA regions that vary between the species. These regions serve as a kind of identifier, yet they also contain regions that are highly conserved, ubiquitous among many species, and are therefore suitable for the PCR. Profiling GIT microbial populations with this technique involves extraction of genomic DNA, which is followed by the use of PCR to amplify that DNA. Amplification utilizes primers targeting conserved, ubiquitous regions of the small subunit rRNA gene, creating what are called “amplicons”. Afterwards, the sequencing of these PCR amplicons takes place.<ref name=Langille></ref> More recently, however, the analysis of the gut microbiome that relies solely on data yielded by NGS has been subject to criticism.<ref name=Jian>[https://doi.org/10.1371/journal.pone.0227285 Jian C, Luukkonen P, Yki-Järvinen H, Salonen A, Korpela K (2020) Quantitative PCR provides a simple and accessible method for quantitative microbiota profiling. PLoS ONE 15(1): e0227285.]</ref> Research indicates that NGS can lead to misinterpretations of microbial community structure, due to the amplification of one taxon possibly leading to the decrease of the other, altering the compositional data.<ref name=Jian></ref> Quantitative PCR (qPCR) has been suggested as an effective addition to the NGS, providing an accurate estimation of absolute taxon abundances in the microbial community.<ref name=Jian></ref> Its benefits over more traditional approaches have been identified as cost-effectiveness, feasibility, simplicity, and applicability.<ref name=Jian></ref> NGS and qPCR both target bacterial DNA and qPCR is expected to reflect the NGS-targeted community structure in a more accurate manner both quantitatively and qualitatively. | ||
===Shotgun Sequencing=== | ===Shotgun Sequencing=== | ||
Shotgun sequencing is a technique used to detect novel genes without having any prior DNA sequences.<ref name=Adams>[https://www.nature.com/scitable/topicpage/complex-genomes-shotgun-sequencing-609/ Jill U. Adams, Ph.D. (Freelance science writer in Albany, NY) © 2008 Nature Education | Shotgun sequencing is a technique used to detect novel genes without having any prior DNA sequences.<ref name=Adams>[https://www.nature.com/scitable/topicpage/complex-genomes-shotgun-sequencing-609/ Jill U. Adams, Ph.D. (Freelance science writer in Albany, NY) © 2008 Nature Education | ||
Citation: Adams, J. (2008) Complex genomes: Shotgun sequencing. Nature Education 1(1):186]</ref> This can be done through the breaking up of the genome into small DNA fragments followed by molecular manipulations of nucleic acids to attach known sequences for primers that will perform sequencing reactions to unknown sequences.<ref name=Adams></ref> | Citation: Adams, J. (2008) Complex genomes: Shotgun sequencing. Nature Education 1(1):186]</ref> This can be done through the breaking up of the genome into small DNA fragments followed by molecular manipulations of nucleic acids to attach known sequences for primers that will perform sequencing reactions to unknown sequences.<ref name=Adams></ref> Shotgun sequencing, therefore, provides a comprehensive view of microbial communities, gene content, and expression levels.<ref name=Adams></ref> This method also has the advantage of simplicity and rapid occurrence, but its limitation is exposed by genomes with numerous repeated regions, since it tends to not work very well with those.<ref name=Adams></ref> These genomes (like the human genome) create difficulties with chromosome assembly.<ref name=Adams></ref> Despite this disadvantage, shotgun sequencing remains an efficacious method that provides valuable insight into identifying active organisms and metabolic activities at the time of sampling and directly links specific functional capabilities to a given microbial lineage.<ref name=Greenblum>[https://doi.org/10.1073/pnas.1116053109 Greenblum, S., Turnbaugh, P. J., & Borenstein, E. (2012). Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proceedings of the National Academy of Sciences of the United States of America, 109(2), 594–599.]</ref> Such a thorough analysis can identify essential GIT microbiota community members as well as vital genes associated with the correct functioning of the GIT microbiota.<ref name=Greenblum></ref> This allows for prediction of the effects various factors may have on the GIT microbiota’s composition and its overall performance.<ref name=Greenblum></ref> | ||
==Alcohol as a Culprit of Dysbiosis in the Intestinal Microbiota== | ==Alcohol as a Culprit of Dysbiosis in the Intestinal Microbiota== | ||
===Changes of the composition of gut microbiota=== | ===Changes of the composition of gut microbiota=== | ||
[[Image:Wiki_research_figure_3.jpg|thumb|500px|left| Bacterial composition of alcohol overconsumer's vs. abstinent's gut environment. https://www.tandfonline.com/doi/full/10.1080/19490976.2019.1580097 ]] | [[Image:Wiki_research_figure_3.jpg|thumb|500px|left| Bacterial composition of an alcohol overconsumer's vs. an abstinent's gut environment. https://www.tandfonline.com/doi/full/10.1080/19490976.2019.1580097 ]] | ||
The most frequent mechanism by which alcohol induces dysbiosis is changing the composition of the gut microbiota.<ref name=Chen>[https://doi.org/10.1002/hep.24423 Chen, Y., Yang, F., Lu, H., Wang, B., Chen, Y., Lei, D., Wang, Y., Zhu, B., & Li, L. (2011). Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology (Baltimore, Md.), 54(2), 562–572.]</ref> This happens because alcohol and its metabolites either inhibit or promote the proliferation of gut bacteria, which significantly alters the intestinal microenvironment.<ref name=Chen>[https://doi.org/10.1002/hep.24423 Chen, Y., Yang, F., Lu, H., Wang, B., Chen, Y., Lei, D., Wang, Y., Zhu, B., & Li, L. (2011). Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology (Baltimore, Md.), 54(2), 562–572.]</ref> This is a key point, as recent research<ref name=Xiao>[https://doi.org/10.1016/j.toxlet.2018.01.021 Xiao, H. W., Ge, C., Feng, G. X., Li, Y., Luo, D., Dong, J. L., Li, H., Wang, H., Cui, M., & Fan, S. J. (2018). Gut microbiota modulates alcohol withdrawal-induced anxiety in mice. Toxicology letters, 287, 23–30.]</ref> proves that the most notable effects of alcohol exposure evolve from changing the composition of the gut microbiota. An experimentally-proven trend of how relative abundance of most bacterial species in the gut microbiome is altered after chronic alcohol overconsumption has been observed. Generally, the presence of Proteobacteria, Enterobacteriaceae, Fusobacteria<ref name=Chen>[https://doi.org/10.1002/hep.24423 Chen, Y., Yang, F., Lu, H., Wang, B., Chen, Y., Lei, D., Wang, Y., Zhu, B., & Li, L. (2011). Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology (Baltimore, Md.), 54(2), 562–572.]</ref> increases. These increased bacteria belong to the family of gram-negative bacteria, suggesting that the alcohol- | The most frequent mechanism by which alcohol induces dysbiosis is changing the composition of the gut microbiota.<ref name=Chen>[https://doi.org/10.1002/hep.24423 Chen, Y., Yang, F., Lu, H., Wang, B., Chen, Y., Lei, D., Wang, Y., Zhu, B., & Li, L. (2011). Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology (Baltimore, Md.), 54(2), 562–572.]</ref> This happens because alcohol and its metabolites either inhibit or promote the proliferation of gut bacteria, which significantly alters the intestinal microenvironment.<ref name=Chen>[https://doi.org/10.1002/hep.24423 Chen, Y., Yang, F., Lu, H., Wang, B., Chen, Y., Lei, D., Wang, Y., Zhu, B., & Li, L. (2011). Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology (Baltimore, Md.), 54(2), 562–572.]</ref> This is a key point, as recent research<ref name=Xiao>[https://doi.org/10.1016/j.toxlet.2018.01.021 Xiao, H. W., Ge, C., Feng, G. X., Li, Y., Luo, D., Dong, J. L., Li, H., Wang, H., Cui, M., & Fan, S. J. (2018). Gut microbiota modulates alcohol withdrawal-induced anxiety in mice. Toxicology letters, 287, 23–30.]</ref> proves that the most notable effects of alcohol exposure evolve from changing the composition of the gut microbiota. An experimentally-proven trend of how relative abundance of most bacterial species in the gut microbiome is altered after chronic alcohol overconsumption has been observed. Generally, the presence of <i>Proteobacteria</i>, <i>Enterobacteriaceae</i>, and <i>Fusobacteria</i><ref name=Chen>[https://doi.org/10.1002/hep.24423 Chen, Y., Yang, F., Lu, H., Wang, B., Chen, Y., Lei, D., Wang, Y., Zhu, B., & Li, L. (2011). Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology (Baltimore, Md.), 54(2), 562–572.]</ref> increases. These increased bacteria belong to the family of gram-negative bacteria, suggesting that the alcohol-exposed gut microbiome is composed of a significantly higher ratio of gram-negative bacteria, which then serves as an ideal environment for lipopolysaccharides (LPS) - induced inflammation.<ref name=Chen>[https://doi.org/10.1002/hep.24423 Chen, Y., Yang, F., Lu, H., Wang, B., Chen, Y., Lei, D., Wang, Y., Zhu, B., & Li, L. (2011). Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology (Baltimore, Md.), 54(2), 562–572.]</ref> On the other hand, there are plenty of bacterial species whose relative abundance in the GIT decreases as a result of alcohol exposure. These bacteria include <i>Lactobacillus</i> and <i>Bifidobacterium</i><ref name=Chen>[https://doi.org/10.1002/hep.24423 Chen, Y., Yang, F., Lu, H., Wang, B., Chen, Y., Lei, D., Wang, Y., Zhu, B., & Li, L. (2011). Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology (Baltimore, Md.), 54(2), 562–572.]</ref>, which reduce alcohol-induced gastrointestinal inflammation and alterations in gut microbiota composition. The presence of <i>Muribaculum intestinale</i>, <i>Ruminococcus</i>, and <i>Faecalibacterium prausnitzi</i><ref name=Smith>[https://doi.org/10.1186/s12866-019-1494-7 Smith, B.J., Miller, R.A., Ericsson, A.C. et al. Changes in the gut microbiome and fermentation products concurrent with enhanced longevity in acarbose-treated mice. BMC Microbiol 19, 130 (2019).]</ref> also decreases. These beneficial bacteria produce short-chain fatty acids (SCFAs), important agents involved in processes of gut function, maintenance of immunity, and inflammation reduction.<ref name=Hamer>[https://doi.org/10.1111/j.1365-2036.2007.03562.x Hamer, H. M., Jonkers, D., Venema, K., Vanhoutvin, S., Troost, F. J., & Brummer, R. J. (2008). Review article: the role of butyrate on colonic function. Alimentary pharmacology & therapeutics, 27(2), 104–119.]</ref> Overall, the compositional changes in alcohol-induced gut composition demonstrate an increase in harmful bacteria and a decrease in beneficial bacteria. | ||
===Changes of the diversity of gut microbiota=== | ===Changes of the diversity of gut microbiota=== | ||
[[Image:Wiki_research_figure_4.jpg|thumb|500px|left| Summary of bacterial species and phyla present in the gut microbiome of alcohol consumer vs abstinent. https://www.tandfonline.com/doi/full/10.1080/19490976.2019.1580097]] | [[Image:Wiki_research_figure_4.jpg|thumb|500px|left| Summary of bacterial species and phyla present in the gut microbiome of an alcohol consumer vs. an abstinent. https://www.tandfonline.com/doi/full/10.1080/19490976.2019.1580097]] | ||
Changes in the diversity of gut microbiome arise from altered levels of downregulation and upregulation of microbiome’s components, rather than its relative abundance.<ref name=Chen>[https://doi.org/10.1002/hep.24423 Chen, Y., Yang, F., Lu, H., Wang, B., Chen, Y., Lei, D., Wang, Y., Zhu, B., & Li, L. (2011). Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology (Baltimore, Md.), 54(2), 562–572.]</ref> Fluctuations in the diversity of the gut microbiota differ based on the drinking patterns | Changes in the diversity of gut microbiome arise from altered levels of downregulation and upregulation of microbiome’s components, rather than its relative abundance.<ref name=Chen>[https://doi.org/10.1002/hep.24423 Chen, Y., Yang, F., Lu, H., Wang, B., Chen, Y., Lei, D., Wang, Y., Zhu, B., & Li, L. (2011). Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology (Baltimore, Md.), 54(2), 562–572.]</ref> Fluctuations in the diversity of the gut microbiota differ based on the drinking patterns<ref name=Chen>[https://doi.org/10.1002/hep.24423 Chen, Y., Yang, F., Lu, H., Wang, B., Chen, Y., Lei, D., Wang, Y., Zhu, B., & Li, L. (2011). Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology (Baltimore, Md.), 54(2), 562–572.]</ref>, which are divided into 2 subgroups - chronic (years of chronic drinking) and acute (recent heavy drinking). | ||
Division of drinking patterns | Division of drinking patterns was not relevant in the previous discussion of compositional changes, as chronic and acute drinking have roughly the same effect in that category. However, it becomes immensely important what drinking pattern is exhibited when investigating the alterations in the diversity of gut microbiota. Changes in gut microbiota caused by acute alcohol exposure are momentary and reversible<ref name=Ming>[https://doi.org/10.3168/jds.2019-17247 Ming, L., Qiao, X., Yi, L., Siren, D., He, J., Hai, L., Guo, F., Xiao, Y., & Ji, R. (2020). Camel milk modulates ethanol-induced changes in the gut microbiome and transcriptome in a mouse model of acute alcoholic liver disease. Journal of dairy science, 103(5), 3937–3949.]</ref>, whereas chronic alcohol exposure has much more severe effects that lead to complete changes in the diversity of gut microbiota.<ref name=Bjørkhaug>[https://doi.org/10.1080/19490976.2019.1580097 Bjørkhaug, S. T., Aanes, H., Neupane, S. P., Bramness, J. G., Malvik, S., Henriksen, C., Skar, V., Medhus, A. W., & Valeur, J. (2019). Characterization of gut microbiota composition and functions in patients with chronic alcohol overconsumption. Gut microbes, 10(6), 663–675.]</ref> This is supported by a recent study<ref name=Lee>[https://doi.org/10.1016/j.foodres.2019.108800 Lee, J. E., Ha, J. S., Park, H. Y., & Lee, E. (2020). Alteration of gut microbiota composition by short-term low-dose alcohol intake is restored by fermented rice liquor in mice. Food research international (Ottawa, Ont.), 128, 108800.]</ref>, which reveals that gut microbiomes affected by different dosages of consumed alcohol will return to the expression of a normal gut environment at varying times. Both short-term and long-term exposure to alcohol upregulates levels of the phyla <i>Firmicutes</i><ref name=Bjørkhaug></ref>, <i>Actinobacteria</i><ref name=Bjørkhaug></ref>, and <i>Proteobacteria</i><ref name=Bjørkhaug></ref> and downregulates the phylum <i>Bacteroidetes</i><ref name=Bjørkhaug></ref>. However, additionally, high-dose and long-term alcohol consumption upregulates levels of the genus <i>Helicobacter</i><ref name=<ref name=Bjørkhaug></ref> and downregulates levels of <i>Lactobacillus</i><ref name=Bjørkhaug></ref>, creating an ideal environment for development of alcohol-induced liver injury.<ref name=Lin>[https://pubs.rsc.org/en/content/articlelanding/2020/FO/C9FO02040C Lin D., Jiang X., Zhao Y., Zhai X., Yang X. (2020). Komagataeibacter hansenii CGMCC 3917 alleviates alcohol-induced liver injury by regulating fatty acid metabolism and intestinal microbiota diversity in mice. Food Funct. 11 4591–4604. 10.1039/c9fo02040c]</ref> Therefore, the disruption of diversity of gut microbiota caused by high-dose and long-term alcohol consumption requires a longer time and more effortful action of modification to recover<ref name=Lin>[https://pubs.rsc.org/en/content/articlelanding/2020/FO/C9FO02040C Lin D., Jiang X., Zhao Y., Zhai X., Yang X. (2020). Komagataeibacter hansenii CGMCC 3917 alleviates alcohol-induced liver injury by regulating fatty acid metabolism and intestinal microbiota diversity in mice. Food Funct. 11 4591–4604. 10.1039/c9fo02040c]</ref>, in comparison to short-term and low-dose alcohol consumption whose effects can be restored in relatively simple ways, such as boosting the diversity of gut microbiome by supplementation of proper probiotics. <ref name=Chen></ref> This substantial difference is likely correlated with the severe effects of higher concentrations of alcohol causing inflammation and liver injury.<ref name=Chen></ref> | ||
==Alcohol-Induced Diseases of the GIT== | ==Alcohol-Induced Diseases of the GIT== | ||
Once the composition and diversity of the gut microbiome has been altered, the entire complex becomes highly susceptible to pathological changes of the system.<ref name=Lozupone>[https://doi.org/10.1038/nature11550 Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K., & Knight, R. (2012). Diversity, stability and resilience of the human gut microbiota. Nature, 489(7415), 220–230.]</ref> The combination of dysbiosis caused by alcohol-induced bacterial overgrowth and disruption of intestinal barrier structure and function is a perfect storm for the development of alcohol-induced liver pathology.<ref name=Engen>[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4590619/ Engen, P. A., Green, S. J., Voigt, R. M., Forsyth, C. B., & Keshavarzian, A. (2015). The Gastrointestinal Microbiome: Alcohol Effects on the Composition of Intestinal Microbiota. Alcohol research : current reviews, 37(2), 223–236.]</ref> Subcategories include nonalcoholic fatty liver disease (NAFLD)<ref name=Engen></ref>, nonalcoholic steatohepatitis (NASH)<ref name=Engen></ref>, and alcohol related liver disease (ALD).<ref name=Engen></ref> The disruption of intestinal barrier, specifically, is a very serious occurrence that has large-scale implications. The catalyst of this event is the oxidative stress in the intestine caused by alcohol consumption and the final product is disruption of tight junctions and promotion of intestinal hyperpermeability.<ref name=Engen></ref> The latter allows for translocation of highly dangerous proinflammatory microbial products from the intestinal lumen to the liver.<ref name=Engen></ref> This is the direct cause of inflammation in the liver<ref name=Engen></ref>, a misfortune many alcoholics experience. Intestinal environment is additionally threatened by alcohol and its metabolites damaging the intestinal mucosa.<ref name=Chen></ref> Alcohol has been recently found to influence MUC-2<ref name=Chen></ref>, the building unit of intestinal mucus layer. By manipulation of MUC- | Once the composition and the diversity of the gut microbiome has been altered, the entire complex becomes highly susceptible to pathological changes of the system.<ref name=Lozupone>[https://doi.org/10.1038/nature11550 Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K., & Knight, R. (2012). Diversity, stability and resilience of the human gut microbiota. Nature, 489(7415), 220–230.]</ref> The combination of dysbiosis caused by alcohol-induced bacterial overgrowth and disruption of intestinal barrier structure and function is a perfect storm for the development of alcohol-induced liver pathology.<ref name=Engen>[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4590619/ Engen, P. A., Green, S. J., Voigt, R. M., Forsyth, C. B., & Keshavarzian, A. (2015). The Gastrointestinal Microbiome: Alcohol Effects on the Composition of Intestinal Microbiota. Alcohol research : current reviews, 37(2), 223–236.]</ref> Subcategories include nonalcoholic fatty liver disease (NAFLD)<ref name=Engen></ref>, nonalcoholic steatohepatitis (NASH)<ref name=Engen></ref>, and alcohol related liver disease (ALD).<ref name=Engen></ref> The disruption of intestinal barrier, specifically, is a very serious occurrence that has large-scale implications. The catalyst of this event is the oxidative stress in the intestine caused by alcohol consumption and the final product is disruption of tight junctions and promotion of intestinal hyperpermeability.<ref name=Engen></ref> The latter allows for translocation of highly dangerous proinflammatory microbial products from the intestinal lumen to the liver.<ref name=Engen></ref> This is the direct cause of inflammation in the liver<ref name=Engen></ref>, a misfortune many alcoholics experience. Intestinal environment is additionally threatened by alcohol and its metabolites damaging the intestinal mucosa.<ref name=Chen></ref> Alcohol has been recently found to influence MUC-2<ref name=Chen></ref>, the building unit of intestinal mucus layer. By manipulation of MUC-2, alcohol metabolites can successfully increase the thickness of this intestinal mucus layer.<ref name=Chen></ref> The specific agent that causes this abnormality is acetaldehyde.<ref name=Chen></ref> Acetaldehyde is an important metabolite involved in alcohol metabolism. Gut microbiota is unable to metabolize acetaldehyde and its build-up causes severe oxidative stress that is involved in altering the levels of expression of MUC-2.<ref name=Chen></ref> Fortunately, this valuable insight into the mechanism of intestinal mucosa damage identifies acetaldehyde as a possible target<ref name=Chen></ref> for treatments involved in repairing alcoholic intestinal mucosal injury. | ||
Human organism functions as a well-oiled machine, requiring careful coordination of many parts. Just one of these parts becoming impaired can have a serious effect on the other parts, and the organism as a whole. That remains true in the case of an impaired gut microbiome, too, as it negatively impacts other systems. | The other predominant disease associated with excessive use of alcohol is cirrhosis<ref name=Lozupone></ref>, the extreme case of alcoholic liver disease. Alcoholic cirrhosis can be characterized by significantly altered intestinal structure and metabolism<ref name=Chen></ref>, resulting in a liver that is swollen, stiff, and barely able to function at any level. Bacteria-wise, cirrhosis expands the presence of <i>Bacteroidetes</i><ref name=Bjørkhaug></ref> in the gut microbiome, as a result of decreasing concentration of butyric acid<ref name=Bjørkhaug></ref> found in patients with cirrhosis. Butyric acid is an essential factor for the gut homeostasis, especially the maintenance of the intestinal wall integrity.<ref name=Bjørkhaug></ref> A high concentration of butyric acid is desired, as it confers protection against the inflammatory activity.<ref name=Bjørkhaug></ref> Since the formation of butyric acid in the gut microbiome depends on gram-positive bacteria <i>Firmicutes</i><ref name=Bjørkhaug></ref>, such as <i>Faecalibacterium prausnitzii</i>, it is predicted that the abundance of this species is minimized because of expanding <i>Bacteroidetes</i> in the gut microbiome.<ref name=Bjørkhaug></ref> | ||
Human organism functions as a well-oiled machine, requiring careful coordination of many parts. Just one of these parts becoming impaired can have a serious effect on the other parts, and the organism as a whole. That remains true in the case of an impaired gut microbiome, too, as it negatively impacts other systems. For example, the brain and the central nervous system (CNS). Generally, alcohol has obvious negative effects on the brain and CNS, but more interestingly, a fascinating study<ref name=Nettleton>[https://doi.org/10.3390/nu11061248 Nettleton, J. E., Klancic, T., Schick, A., Choo, A. C., Shearer, J., Borgland, S. L., Chleilat, F., Mayengbam, S., & Reimer, R. A. (2019). Low-Dose Stevia (Rebaudioside A) Consumption Perturbs Gut Microbiota and the Mesolimbic Dopamine Reward System. Nutrients, 11(6), 1248.]</ref> found correlations between CNS, dopamine levels, and gut dysbiosis. Release of dopamine is mediated via careful balance between its concentration and dopamine receptors of the CNS.<ref name=Volkow>[https://doi.org/10.1021/acschemneuro.5b00067 Volkow, N. D., Koob, G., & Baler, R. (2015). Biomarkers in substance use disorders. ACS chemical neuroscience, 6(4), 522–525.]</ref> After alcohol enters the human body its metabolites disrupt this homeostasis by changing the ratio of ligands and linking receptors<ref name=Volkow></ref>, causing the dysfunction of the CNS. Furthermore, this imbalance is enhanced by gut microbiome dysbiosis<ref name=Nettleton></ref>, which decreases the level of dopamine even more. This most likely happens due to the gut microbiome trying to mediate its own metabolites<ref name=Nettleton></ref> in the presence of alcohol metabolites, an energetically-expensive task. Its indirect result is downregulating the expression of important dopamine transporter genes<ref name=Nettleton></ref>, decreasing the levels of dopamine in the brain. | |||
== Possible Treatments and Therapeutic Interventions== | == Possible Treatments and Therapeutic Interventions== | ||
[[Image:Wiki_lactobacillus_alcohol.PNG|thumb|300px|right| | [[Image:Wiki_lactobacillus_alcohol.PNG|thumb|300px|right| <i>Lactobacillus plantarum</i>, beneficial bacteria that provides relief and restoration of the gut microbiome damaged by alcohol use. https://neurosciencenews.com/neuroscience-terms/lactobacillus-plantarum/ ]] | ||
The debate on whether alcohol-induced gut microbiome dysbiosis and its health consequences may be reversible, and if so, how, has been a heated debate in science circles the past few years.<ref name=Engen></ref> Probiotic<ref name=Engen></ref> and synbiotic<ref name=Engen></ref> interventions might be a promising solution. Probiotics are live microorganisms that maintain the normal microflora in the body by expanding abundance of essential bacteria of the gut<ref name=Pandey>[https://doi.org/10.1007/s13197-015-1921-1 Pandey, K. R., Naik, S. R., & Vakil, B. V. (2015). Probiotics, prebiotics and synbiotics- a review. Journal of food science and technology, 52(12), 7577–7587.]</ref> Synbiotics are a combination of probiotics and prebiotics that improve the functioning of aforementioned essential bacteria, contributing to the overall well-being of the host<ref name=Pandey></ref>. There are plenty of bacteria that significantly attenuate gastrointestinal inflammation by reducing activity of myeloperoxidase<ref name=Chen></ref>, an agent linked to inflammatory reactions. Bacteria such as is Lactobacillus Plantarum<ref name=Chen></ref>, commonly found in many fermented food products. Studies on this bacterium have shown that Lactobacillus Plantarum can regulate the intestinal barrier damaged by alcohol overconsumption in a fashion that provides relief, but not riddance of liver injury.<ref name=Chen></ref> Yet another study<ref name=Han>[https://doi.org/10.1080/19490976.2018.1479625 Han, X., Lee, A., Huang, S., Gao, J., Spence, J. R., & Owyang, C. (2019). Lactobacillus rhamnosus GG prevents epithelial barrier dysfunction induced by interferon-gamma and fecal supernatants from irritable bowel syndrome patients in human intestinal enteroids and colonoids. Gut microbes, 10(1), 59–76.]</ref> reveals that this bacteria is remarkably effective with not only liver injury, but also irritable bowel syndrome (IBS).<ref name=Han></ref> This is an important finding as symptoms of both occur concurrently in most alcoholic patients.<ref name=Han></ref> | The debate on whether alcohol-induced gut microbiome dysbiosis and its health consequences may be reversible, and if so, how, has been a heated debate in science circles the past few years.<ref name=Engen></ref> Probiotic<ref name=Engen></ref> and synbiotic<ref name=Engen></ref> interventions might be a promising solution. Probiotics are live microorganisms that maintain the normal microflora in the body by expanding abundance of essential bacteria of the gut.<ref name=Pandey>[https://doi.org/10.1007/s13197-015-1921-1 Pandey, K. R., Naik, S. R., & Vakil, B. V. (2015). Probiotics, prebiotics and synbiotics- a review. Journal of food science and technology, 52(12), 7577–7587.]</ref> Synbiotics are a combination of probiotics and prebiotics that improve the functioning of aforementioned essential bacteria, contributing to the overall well-being of the host<ref name=Pandey></ref>. There are plenty of bacteria that significantly attenuate gastrointestinal inflammation by reducing activity of myeloperoxidase<ref name=Chen></ref>, an agent linked to inflammatory reactions. Bacteria such as is <i>Lactobacillus Plantarum</i><ref name=Chen></ref>, commonly found in many fermented food products. Studies on this bacterium have shown that <i>Lactobacillus Plantarum</i> can regulate the intestinal barrier damaged by alcohol overconsumption in a fashion that provides relief, but not a complete riddance of liver injury.<ref name=Chen></ref> Yet another study<ref name=Han>[https://doi.org/10.1080/19490976.2018.1479625 Han, X., Lee, A., Huang, S., Gao, J., Spence, J. R., & Owyang, C. (2019). Lactobacillus rhamnosus GG prevents epithelial barrier dysfunction induced by interferon-gamma and fecal supernatants from irritable bowel syndrome patients in human intestinal enteroids and colonoids. Gut microbes, 10(1), 59–76.]</ref> reveals that this bacteria is remarkably effective with not only liver injury, but also irritable bowel syndrome (IBS).<ref name=Han></ref> This is an important finding as symptoms of both occur concurrently in most alcoholic patients.<ref name=Han></ref> | ||
Other popular form of treatment are corticosteroids.<ref name=Osna>[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5513682/ Osna, N. A., Donohue, T. M., Jr, & Kharbanda, K. K. (2017). Alcoholic Liver Disease: Pathogenesis and Current Management. Alcohol research : current reviews, 38(2), 147–161.]</ref> These include the use of prednisolone<ref name=Osna></ref>, a highly efficacious anti-inflammatory agent. Its ability to suppress proinflammatory cytokine response of the immune system<ref name=Osna></ref> induces early improvement in liver function<ref name=Mathurin></ref> of a patient with moderate to severe alcoholic hepatitis. This contributes to short-term<ref name=Mathurin>[https://doi.org/10.1001/jama.2013.276300 Mathurin, P., Louvet, A., Duhamel, A., Nahon, P., Carbonell, N., Boursier, J., Anty, R., Diaz, E., Thabut, D., Moirand, R., Lebrec, D., Moreno, C., Talbodec, N., Paupard, T., Naveau, S., Silvain, C., Pageaux, G. P., Sobesky, R., Canva-Delcambre, V., Dharancy, S., … Dao, T. (2013). Prednisolone with vs without pentoxifylline and survival of patients with severe alcoholic hepatitis: a randomized clinical trial. JAMA, 310(10), 1033–1041.]</ref> effects in healing alcoholic hepatitis. More research is needed to establish how successful prednisolone is in treating the disease for good.<ref name=Mathurin></ref> | Other popular form of treatment are corticosteroids.<ref name=Osna>[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5513682/ Osna, N. A., Donohue, T. M., Jr, & Kharbanda, K. K. (2017). Alcoholic Liver Disease: Pathogenesis and Current Management. Alcohol research : current reviews, 38(2), 147–161.]</ref> These include the use of prednisolone<ref name=Osna></ref>, a highly efficacious anti-inflammatory agent. Its ability to suppress proinflammatory cytokine response of the immune system<ref name=Osna></ref> induces early improvement in liver function<ref name=Mathurin></ref> of a patient with moderate to severe alcoholic hepatitis. This contributes to short-term<ref name=Mathurin>[https://doi.org/10.1001/jama.2013.276300 Mathurin, P., Louvet, A., Duhamel, A., Nahon, P., Carbonell, N., Boursier, J., Anty, R., Diaz, E., Thabut, D., Moirand, R., Lebrec, D., Moreno, C., Talbodec, N., Paupard, T., Naveau, S., Silvain, C., Pageaux, G. P., Sobesky, R., Canva-Delcambre, V., Dharancy, S., … Dao, T. (2013). Prednisolone with vs without pentoxifylline and survival of patients with severe alcoholic hepatitis: a randomized clinical trial. JAMA, 310(10), 1033–1041.]</ref> effects in healing alcoholic hepatitis. More research is needed to establish how successful prednisolone is in treating the disease for good.<ref name=Mathurin></ref> | ||
Latest revision as of 15:59, 1 May 2023
Introduction
By Nikola Kovacova
The gut microbiota is classified as a collection of all microbial organisms within the gastrointestinal tract (GIT).[1] With trillions of microorganisms inhabiting the microbiota, its collective genome is believed to encode 100 times more genes than the human genome.[2] This makes it not only a complex, but also an essential player involved in the health status of the GIT, as it impacts digestion, inflammation, and immunity.[3] The microbiota–host relationship is mutualistic - the microbiota contributes to the extraction of energy from various sources, boosts synthesis of vitamins and amino acids, and assists with the formation of barriers against pathogens.[4] This relationship, however, is highly sensitive towards imbalances in microbial composition that can often lead to a decrease in overall well-being of the host.[5] This imbalance, referred to as dysbiosis, can be easily caused by environmental factors incorporated into the lives of countless people in the contemporary Western societies, especially the poor dietary choices. Diets high in fat and sugar have serious negative consequences on the microbiota composition and diversity, contributing to the development of obesity and liver injury, as well as multiple diseases such as IBD, IBS, celiac disease, type 1 and type 2 diabetes, food allergies, and cardiovascular disease.[5]
This page focuses on one specific dietary disruptor of the gut microbiota. Alcohol. Alcoholism is a worldwide problem with implications far beyond the healthcare sector. Drastic social, financial, and interpersonal consequences in the life of the person stricken with alcoholism often occur. The World Health Organization [1] has revealed that excessive alcohol consumption is responsible for 3 million deaths yearly and over astonishing 280 million people globally have a diagnosed alcohol use disorder. It has been well-established that the toxicity of the alcohol itself is associated with the formation of various health complications. These might be tissue injury and organ dysfunction, higher risk of developing cancer, or abnormal function of the immune system that increases the risk of acute and chronic infections.[6] This review is interested in addressing alcohol’s specific impact on the microbiota in the gastrointestinal tract (GIT), the compositional and functional changes it induces, and corresponding health-related issues.
How to Analyze the Intestinal Microbial Population?
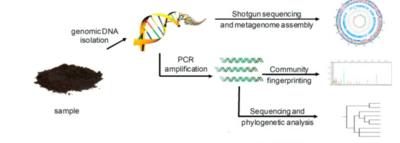
Before we dive into the specific alcohol-induced changes in the microbiota, we must first consider how researchers can evaluate the number of microorganisms in the microbial populations and monitor changes in its size and composition. This is not exactly an easy thing to do, as directly measuring microbial communities such as those within the GIT comes along with certain challenges and ambiguities. Firstly, microorganisms in the gut environment maintain unbelievable genetic diversity but store this diversity in a very limited array of cellular morphologies.[7] Additionally, microorganisms have unessential functions that complicate the analysis, share some of the functional capabilities with closely related microorganisms, and most importantly - are extraordinarily difficult to isolate under laboratory conditions.[7] Consideration of all these factors leads to strong preference towards the use of molecular tools - tools that look for and examine DNA and RNA. This allows for careful analysis of these complex microbial communities. Most commonly, the use of one of the two most popular techniques is utilized. These are polymerase chain reaction (PCR) and shotgun sequencing.
PCR and Total Bacterial Quantification
Technique of PCR consists of walking on a fine line between finding an appropriate gene target, a section that would be frequent enough among microorganisms so that it could be used for searching, but different enough so that it could individuate among microorganisms. Traditionally, utilization of ribosomal RNA (rRNA) genes has been preferred for this molecular analysis. This analysis consists of using PCR-based techniques coupled with indirect fingerprinting or direct sequencing, such as next-generation sequencing (NGS).[8] This is due to rRNA genes containing very specific DNA regions that vary between the species. These regions serve as a kind of identifier, yet they also contain regions that are highly conserved, ubiquitous among many species, and are therefore suitable for the PCR. Profiling GIT microbial populations with this technique involves extraction of genomic DNA, which is followed by the use of PCR to amplify that DNA. Amplification utilizes primers targeting conserved, ubiquitous regions of the small subunit rRNA gene, creating what are called “amplicons”. Afterwards, the sequencing of these PCR amplicons takes place.[8] More recently, however, the analysis of the gut microbiome that relies solely on data yielded by NGS has been subject to criticism.[9] Research indicates that NGS can lead to misinterpretations of microbial community structure, due to the amplification of one taxon possibly leading to the decrease of the other, altering the compositional data.[9] Quantitative PCR (qPCR) has been suggested as an effective addition to the NGS, providing an accurate estimation of absolute taxon abundances in the microbial community.[9] Its benefits over more traditional approaches have been identified as cost-effectiveness, feasibility, simplicity, and applicability.[9] NGS and qPCR both target bacterial DNA and qPCR is expected to reflect the NGS-targeted community structure in a more accurate manner both quantitatively and qualitatively.
Shotgun Sequencing
Shotgun sequencing is a technique used to detect novel genes without having any prior DNA sequences.[10] This can be done through the breaking up of the genome into small DNA fragments followed by molecular manipulations of nucleic acids to attach known sequences for primers that will perform sequencing reactions to unknown sequences.[10] Shotgun sequencing, therefore, provides a comprehensive view of microbial communities, gene content, and expression levels.[10] This method also has the advantage of simplicity and rapid occurrence, but its limitation is exposed by genomes with numerous repeated regions, since it tends to not work very well with those.[10] These genomes (like the human genome) create difficulties with chromosome assembly.[10] Despite this disadvantage, shotgun sequencing remains an efficacious method that provides valuable insight into identifying active organisms and metabolic activities at the time of sampling and directly links specific functional capabilities to a given microbial lineage.[11] Such a thorough analysis can identify essential GIT microbiota community members as well as vital genes associated with the correct functioning of the GIT microbiota.[11] This allows for prediction of the effects various factors may have on the GIT microbiota’s composition and its overall performance.[11]
Alcohol as a Culprit of Dysbiosis in the Intestinal Microbiota
Changes of the composition of gut microbiota
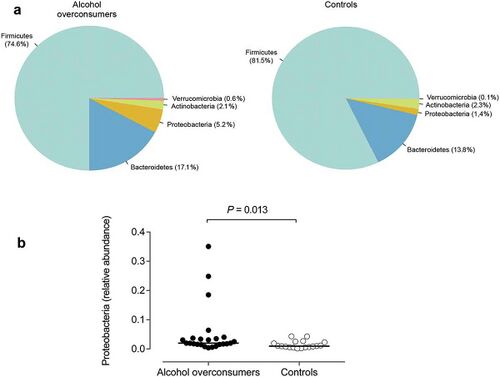
The most frequent mechanism by which alcohol induces dysbiosis is changing the composition of the gut microbiota.[12] This happens because alcohol and its metabolites either inhibit or promote the proliferation of gut bacteria, which significantly alters the intestinal microenvironment.[12] This is a key point, as recent research[13] proves that the most notable effects of alcohol exposure evolve from changing the composition of the gut microbiota. An experimentally-proven trend of how relative abundance of most bacterial species in the gut microbiome is altered after chronic alcohol overconsumption has been observed. Generally, the presence of Proteobacteria, Enterobacteriaceae, and Fusobacteria[12] increases. These increased bacteria belong to the family of gram-negative bacteria, suggesting that the alcohol-exposed gut microbiome is composed of a significantly higher ratio of gram-negative bacteria, which then serves as an ideal environment for lipopolysaccharides (LPS) - induced inflammation.[12] On the other hand, there are plenty of bacterial species whose relative abundance in the GIT decreases as a result of alcohol exposure. These bacteria include Lactobacillus and Bifidobacterium[12], which reduce alcohol-induced gastrointestinal inflammation and alterations in gut microbiota composition. The presence of Muribaculum intestinale, Ruminococcus, and Faecalibacterium prausnitzi[14] also decreases. These beneficial bacteria produce short-chain fatty acids (SCFAs), important agents involved in processes of gut function, maintenance of immunity, and inflammation reduction.[15] Overall, the compositional changes in alcohol-induced gut composition demonstrate an increase in harmful bacteria and a decrease in beneficial bacteria.
Changes of the diversity of gut microbiota
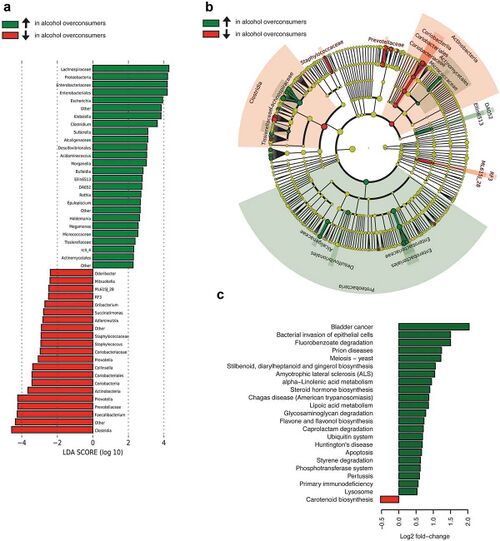
Changes in the diversity of gut microbiome arise from altered levels of downregulation and upregulation of microbiome’s components, rather than its relative abundance.[12] Fluctuations in the diversity of the gut microbiota differ based on the drinking patterns[12], which are divided into 2 subgroups - chronic (years of chronic drinking) and acute (recent heavy drinking). Division of drinking patterns was not relevant in the previous discussion of compositional changes, as chronic and acute drinking have roughly the same effect in that category. However, it becomes immensely important what drinking pattern is exhibited when investigating the alterations in the diversity of gut microbiota. Changes in gut microbiota caused by acute alcohol exposure are momentary and reversible[16], whereas chronic alcohol exposure has much more severe effects that lead to complete changes in the diversity of gut microbiota.[17] This is supported by a recent study[18], which reveals that gut microbiomes affected by different dosages of consumed alcohol will return to the expression of a normal gut environment at varying times. Both short-term and long-term exposure to alcohol upregulates levels of the phyla Firmicutes[17], Actinobacteria[17], and Proteobacteria[17] and downregulates the phylum Bacteroidetes[17]. However, additionally, high-dose and long-term alcohol consumption upregulates levels of the genus Helicobacter[17] and downregulates levels of Lactobacillus[17], creating an ideal environment for development of alcohol-induced liver injury.[19] Therefore, the disruption of diversity of gut microbiota caused by high-dose and long-term alcohol consumption requires a longer time and more effortful action of modification to recover[19], in comparison to short-term and low-dose alcohol consumption whose effects can be restored in relatively simple ways, such as boosting the diversity of gut microbiome by supplementation of proper probiotics. [12] This substantial difference is likely correlated with the severe effects of higher concentrations of alcohol causing inflammation and liver injury.[12]
Alcohol-Induced Diseases of the GIT
Once the composition and the diversity of the gut microbiome has been altered, the entire complex becomes highly susceptible to pathological changes of the system.[20] The combination of dysbiosis caused by alcohol-induced bacterial overgrowth and disruption of intestinal barrier structure and function is a perfect storm for the development of alcohol-induced liver pathology.[21] Subcategories include nonalcoholic fatty liver disease (NAFLD)[21], nonalcoholic steatohepatitis (NASH)[21], and alcohol related liver disease (ALD).[21] The disruption of intestinal barrier, specifically, is a very serious occurrence that has large-scale implications. The catalyst of this event is the oxidative stress in the intestine caused by alcohol consumption and the final product is disruption of tight junctions and promotion of intestinal hyperpermeability.[21] The latter allows for translocation of highly dangerous proinflammatory microbial products from the intestinal lumen to the liver.[21] This is the direct cause of inflammation in the liver[21], a misfortune many alcoholics experience. Intestinal environment is additionally threatened by alcohol and its metabolites damaging the intestinal mucosa.[12] Alcohol has been recently found to influence MUC-2[12], the building unit of intestinal mucus layer. By manipulation of MUC-2, alcohol metabolites can successfully increase the thickness of this intestinal mucus layer.[12] The specific agent that causes this abnormality is acetaldehyde.[12] Acetaldehyde is an important metabolite involved in alcohol metabolism. Gut microbiota is unable to metabolize acetaldehyde and its build-up causes severe oxidative stress that is involved in altering the levels of expression of MUC-2.[12] Fortunately, this valuable insight into the mechanism of intestinal mucosa damage identifies acetaldehyde as a possible target[12] for treatments involved in repairing alcoholic intestinal mucosal injury.
The other predominant disease associated with excessive use of alcohol is cirrhosis[20], the extreme case of alcoholic liver disease. Alcoholic cirrhosis can be characterized by significantly altered intestinal structure and metabolism[12], resulting in a liver that is swollen, stiff, and barely able to function at any level. Bacteria-wise, cirrhosis expands the presence of Bacteroidetes[17] in the gut microbiome, as a result of decreasing concentration of butyric acid[17] found in patients with cirrhosis. Butyric acid is an essential factor for the gut homeostasis, especially the maintenance of the intestinal wall integrity.[17] A high concentration of butyric acid is desired, as it confers protection against the inflammatory activity.[17] Since the formation of butyric acid in the gut microbiome depends on gram-positive bacteria Firmicutes[17], such as Faecalibacterium prausnitzii, it is predicted that the abundance of this species is minimized because of expanding Bacteroidetes in the gut microbiome.[17]
Human organism functions as a well-oiled machine, requiring careful coordination of many parts. Just one of these parts becoming impaired can have a serious effect on the other parts, and the organism as a whole. That remains true in the case of an impaired gut microbiome, too, as it negatively impacts other systems. For example, the brain and the central nervous system (CNS). Generally, alcohol has obvious negative effects on the brain and CNS, but more interestingly, a fascinating study[22] found correlations between CNS, dopamine levels, and gut dysbiosis. Release of dopamine is mediated via careful balance between its concentration and dopamine receptors of the CNS.[23] After alcohol enters the human body its metabolites disrupt this homeostasis by changing the ratio of ligands and linking receptors[23], causing the dysfunction of the CNS. Furthermore, this imbalance is enhanced by gut microbiome dysbiosis[22], which decreases the level of dopamine even more. This most likely happens due to the gut microbiome trying to mediate its own metabolites[22] in the presence of alcohol metabolites, an energetically-expensive task. Its indirect result is downregulating the expression of important dopamine transporter genes[22], decreasing the levels of dopamine in the brain.
Possible Treatments and Therapeutic Interventions
The debate on whether alcohol-induced gut microbiome dysbiosis and its health consequences may be reversible, and if so, how, has been a heated debate in science circles the past few years.[21] Probiotic[21] and synbiotic[21] interventions might be a promising solution. Probiotics are live microorganisms that maintain the normal microflora in the body by expanding abundance of essential bacteria of the gut.[24] Synbiotics are a combination of probiotics and prebiotics that improve the functioning of aforementioned essential bacteria, contributing to the overall well-being of the host[24]. There are plenty of bacteria that significantly attenuate gastrointestinal inflammation by reducing activity of myeloperoxidase[12], an agent linked to inflammatory reactions. Bacteria such as is Lactobacillus Plantarum[12], commonly found in many fermented food products. Studies on this bacterium have shown that Lactobacillus Plantarum can regulate the intestinal barrier damaged by alcohol overconsumption in a fashion that provides relief, but not a complete riddance of liver injury.[12] Yet another study[25] reveals that this bacteria is remarkably effective with not only liver injury, but also irritable bowel syndrome (IBS).[25] This is an important finding as symptoms of both occur concurrently in most alcoholic patients.[25]
Other popular form of treatment are corticosteroids.[26] These include the use of prednisolone[26], a highly efficacious anti-inflammatory agent. Its ability to suppress proinflammatory cytokine response of the immune system[26] induces early improvement in liver function[27] of a patient with moderate to severe alcoholic hepatitis. This contributes to short-term[27] effects in healing alcoholic hepatitis. More research is needed to establish how successful prednisolone is in treating the disease for good.[27] A treatment strategy that gets rid of severe alcoholic hepatitis irreversibly is liver transplantation.[26] However, there are some notable obstacles a patient has to overcome to be considered suitable for this transplantation. Firstly, the possibility of a continued alcohol consumption after the surgery is always a huge factor that makes it very complicated for alcoholic patients to receive a new liver.[26] Majority of the patients must demonstrate abstinence from alcohol for 6 months prior to being considered for liver transplantation.[26] Secondly, abstinence from alcohol itself leads to an improvement in liver function in many cases.[26] Long-term abstinence from alcohol relieves alcohol steatosis[26], the enlargement of the liver due to accumulation of macrovesicular fat into large droplets of triglyceride.[26] This has been demonstrated to increase survival rate in cirrhotic patients notably.[26] This positive effect of abstinence can be further enhanced in combination with lifestyle and dietary modifications.[26]
Conclusion
The excessive use of alcohol is a widespread global problem with severe sociological, economic, and health-related consequences. This page summarizes the manifestations of the last-mentioned, with emphasis on the effects on the gut microbiota. The page discusses methods of analysis of the gut microbiota, alcohol-induced alterations of the composition and diversity of the gut environment, development of diseases as a direct outcome of those, and possible treatments to reverse them. Research identifies the increased ratio of gram-negative to gram-positive bacteria in the gut microbiome due to alcohol overconsumption as a driving force of dysbiosis and inflammation. The elevation of susceptibility of the gut towards diseases follows, liver injury and cirrhosis being the most prominent. Treatment is possible, but must be effortful on the patient’s side. Abstinence from alcohol is required for restoration of the normal gut microflora. Normal functioning of the gut can be further enhanced by supplementary use of probiotics.
References
- ↑ Savage, D.C. "Microbial ecology of the gastrointestinal tract." Annual review of microbiology, 31, 107–133.
- ↑ Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., Nielsen, T., Pons, N., Levenez, F., Yamada, T., Mende, D. R., Li, J., Xu, J., Li, S., Li, D., Cao, J., Wang, B., Liang, H., Zheng, H., Xie, Y., … Wang, J. (2010). "A human gut microbial gene catalogue established by metagenomic sequencing." Nature, 464(7285), 59–65.
- ↑ Leclercq, S., de Timary, P., Delzenne, N. M., & Stärkel, P. (2017). "The link between inflammation, bugs, the intestine and the brain in alcohol dependence." Translational psychiatry, 7(2), e1048.
- ↑ Tappenden, K. A., & Deutsch, A. S. (2007). "The physiological relevance of the intestinal microbiota--contributions to human health." Journal of the American College of Nutrition, 26(6), 679S–83S.
- ↑ 5.0 5.1 Kim, B. S., Jeon, Y. S., & Chun, J. (2013). "Current status and future promise of the human microbiome." Pediatric gastroenterology, hepatology & nutrition, 16(2), 71–79.
- ↑ Szabo, G., & Mandrekar, P. (2009). A recent perspective on alcohol, immunity, and host defense. Alcoholism, clinical and experimental research, 33(2), 220–232.
- ↑ 7.0 7.1 Woese C. R. (1987). Bacterial evolution. Microbiological reviews, 51(2), 221–271.
- ↑ 8.0 8.1 Langille, M., Zaneveld, J., Caporaso, J. et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31, 814–821 (2013).
- ↑ 9.0 9.1 9.2 9.3 Jian C, Luukkonen P, Yki-Järvinen H, Salonen A, Korpela K (2020) Quantitative PCR provides a simple and accessible method for quantitative microbiota profiling. PLoS ONE 15(1): e0227285.
- ↑ 10.0 10.1 10.2 10.3 10.4 [https://www.nature.com/scitable/topicpage/complex-genomes-shotgun-sequencing-609/ Jill U. Adams, Ph.D. (Freelance science writer in Albany, NY) © 2008 Nature Education Citation: Adams, J. (2008) Complex genomes: Shotgun sequencing. Nature Education 1(1):186]
- ↑ 11.0 11.1 11.2 Greenblum, S., Turnbaugh, P. J., & Borenstein, E. (2012). Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proceedings of the National Academy of Sciences of the United States of America, 109(2), 594–599.
- ↑ 12.00 12.01 12.02 12.03 12.04 12.05 12.06 12.07 12.08 12.09 12.10 12.11 12.12 12.13 12.14 12.15 12.16 12.17 12.18 Chen, Y., Yang, F., Lu, H., Wang, B., Chen, Y., Lei, D., Wang, Y., Zhu, B., & Li, L. (2011). Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology (Baltimore, Md.), 54(2), 562–572.
- ↑ Xiao, H. W., Ge, C., Feng, G. X., Li, Y., Luo, D., Dong, J. L., Li, H., Wang, H., Cui, M., & Fan, S. J. (2018). Gut microbiota modulates alcohol withdrawal-induced anxiety in mice. Toxicology letters, 287, 23–30.
- ↑ Smith, B.J., Miller, R.A., Ericsson, A.C. et al. Changes in the gut microbiome and fermentation products concurrent with enhanced longevity in acarbose-treated mice. BMC Microbiol 19, 130 (2019).
- ↑ Hamer, H. M., Jonkers, D., Venema, K., Vanhoutvin, S., Troost, F. J., & Brummer, R. J. (2008). Review article: the role of butyrate on colonic function. Alimentary pharmacology & therapeutics, 27(2), 104–119.
- ↑ Ming, L., Qiao, X., Yi, L., Siren, D., He, J., Hai, L., Guo, F., Xiao, Y., & Ji, R. (2020). Camel milk modulates ethanol-induced changes in the gut microbiome and transcriptome in a mouse model of acute alcoholic liver disease. Journal of dairy science, 103(5), 3937–3949.
- ↑ 17.00 17.01 17.02 17.03 17.04 17.05 17.06 17.07 17.08 17.09 17.10 17.11 17.12 Bjørkhaug, S. T., Aanes, H., Neupane, S. P., Bramness, J. G., Malvik, S., Henriksen, C., Skar, V., Medhus, A. W., & Valeur, J. (2019). Characterization of gut microbiota composition and functions in patients with chronic alcohol overconsumption. Gut microbes, 10(6), 663–675.
- ↑ Lee, J. E., Ha, J. S., Park, H. Y., & Lee, E. (2020). Alteration of gut microbiota composition by short-term low-dose alcohol intake is restored by fermented rice liquor in mice. Food research international (Ottawa, Ont.), 128, 108800.
- ↑ 19.0 19.1 Lin D., Jiang X., Zhao Y., Zhai X., Yang X. (2020). Komagataeibacter hansenii CGMCC 3917 alleviates alcohol-induced liver injury by regulating fatty acid metabolism and intestinal microbiota diversity in mice. Food Funct. 11 4591–4604. 10.1039/c9fo02040c
- ↑ 20.0 20.1 Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K., & Knight, R. (2012). Diversity, stability and resilience of the human gut microbiota. Nature, 489(7415), 220–230.
- ↑ 21.0 21.1 21.2 21.3 21.4 21.5 21.6 21.7 21.8 21.9 Engen, P. A., Green, S. J., Voigt, R. M., Forsyth, C. B., & Keshavarzian, A. (2015). The Gastrointestinal Microbiome: Alcohol Effects on the Composition of Intestinal Microbiota. Alcohol research : current reviews, 37(2), 223–236.
- ↑ 22.0 22.1 22.2 22.3 Nettleton, J. E., Klancic, T., Schick, A., Choo, A. C., Shearer, J., Borgland, S. L., Chleilat, F., Mayengbam, S., & Reimer, R. A. (2019). Low-Dose Stevia (Rebaudioside A) Consumption Perturbs Gut Microbiota and the Mesolimbic Dopamine Reward System. Nutrients, 11(6), 1248.
- ↑ 23.0 23.1 Volkow, N. D., Koob, G., & Baler, R. (2015). Biomarkers in substance use disorders. ACS chemical neuroscience, 6(4), 522–525.
- ↑ 24.0 24.1 Pandey, K. R., Naik, S. R., & Vakil, B. V. (2015). Probiotics, prebiotics and synbiotics- a review. Journal of food science and technology, 52(12), 7577–7587.
- ↑ 25.0 25.1 25.2 Han, X., Lee, A., Huang, S., Gao, J., Spence, J. R., & Owyang, C. (2019). Lactobacillus rhamnosus GG prevents epithelial barrier dysfunction induced by interferon-gamma and fecal supernatants from irritable bowel syndrome patients in human intestinal enteroids and colonoids. Gut microbes, 10(1), 59–76.
- ↑ 26.00 26.01 26.02 26.03 26.04 26.05 26.06 26.07 26.08 26.09 26.10 Osna, N. A., Donohue, T. M., Jr, & Kharbanda, K. K. (2017). Alcoholic Liver Disease: Pathogenesis and Current Management. Alcohol research : current reviews, 38(2), 147–161.
- ↑ 27.0 27.1 27.2 Mathurin, P., Louvet, A., Duhamel, A., Nahon, P., Carbonell, N., Boursier, J., Anty, R., Diaz, E., Thabut, D., Moirand, R., Lebrec, D., Moreno, C., Talbodec, N., Paupard, T., Naveau, S., Silvain, C., Pageaux, G. P., Sobesky, R., Canva-Delcambre, V., Dharancy, S., … Dao, T. (2013). Prednisolone with vs without pentoxifylline and survival of patients with severe alcoholic hepatitis: a randomized clinical trial. JAMA, 310(10), 1033–1041.
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2023, Kenyon College
