Methylothermus thermalis: Difference between revisions
No edit summary |
No edit summary |
||
| Line 37: | Line 37: | ||
<br> | <br> | ||
<br><br> | <br><br> | ||
<br> | |||
<br><br> | |||
<br> | |||
<br><br> | |||
<br> | |||
<br><br> | |||
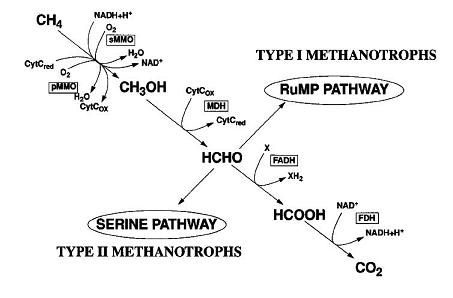
[[Image:Metabolism_type.jpg|thumb|550px|right| Pathways for the oxidation of methane and assimilation of formaldehyde. Abbreviations: CytC, cytochrome c; FADH, formaldehyde dehydrogenase; FDH,formate dehydrogenase. From https://www.ocean.udel.edu/cms/thanson/TH-MicroRev.pdf Hanson R and Hanson T, 1996.].]] | [[Image:Metabolism_type.jpg|thumb|550px|right| Pathways for the oxidation of methane and assimilation of formaldehyde. Abbreviations: CytC, cytochrome c; FADH, formaldehyde dehydrogenase; FDH,formate dehydrogenase. From https://www.ocean.udel.edu/cms/thanson/TH-MicroRev.pdf Hanson R and Hanson T, 1996.].]] | ||
Revision as of 06:26, 21 April 2008
Classification
Bacteria; Proteobacteria; Gammaproteobacteria; Methylococcales; Methylococcaceae
Species
|
Methylothermus thermalis |
|
NCBI: Taxonomy |
Description and Significance
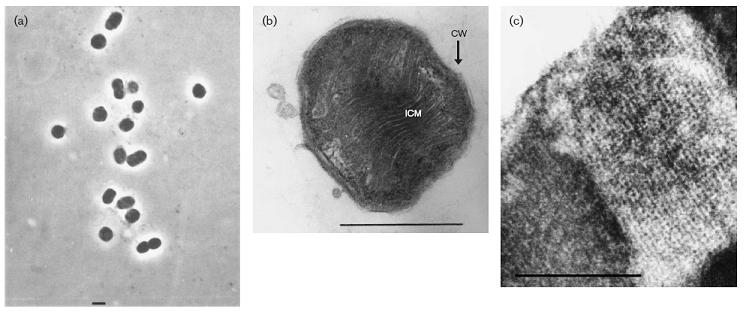
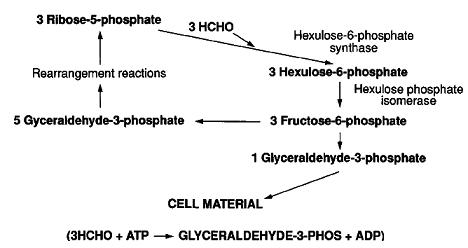
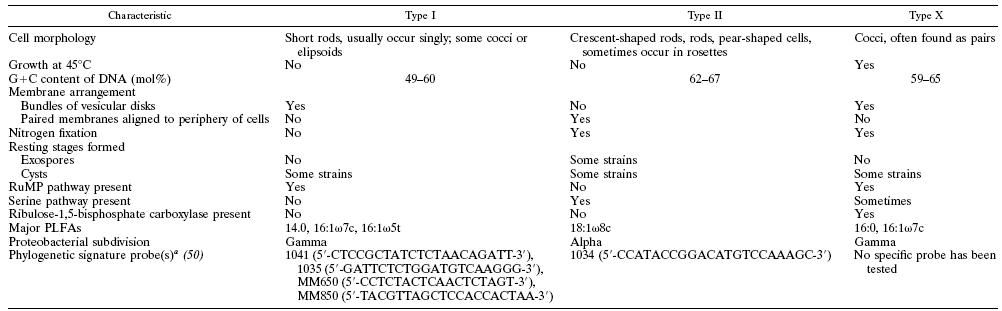
This organism was found in a Japanese hot spring (location not specified). Methylothermus thermalis grew as small (1–2 mm in diameter), white, semi-transparent colonies with an entire edge and smooth surface were obsevered after 1-2 weeks. Older colonies (1 month) were light brown and more rigid. Unlike most discovered methanotrophs, this species can survive at high temperatures while metabolizing methane and methanol. It is classified as a type I thermophilic methanotroph, which uses the RuMP pathway instead of the type II serine pathway.
Genome Structure
The nifH gene, which codes for nitrogenase reductase, was not detected with PCR analysis. Since nitrogenase reductase is component II of the nitrogenase enzyme complex, M. thermalis cannot fix its own atmospheric nitrogen. Also, the mmoX gene was not detected by PCR using the respective primer set that is universal for the known mmoX genes of methanotrophs. The genome of this microorganism has yet to be sequenced and as a consequence the size and characteristics are not well known.
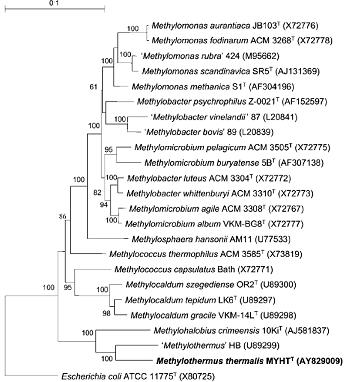
The G+C DNA content of 62.5 mol% for M. thermalis is close to that found for type II and type X methanotrophs. The 16S rRNA gene database search showed that this organism is closely related to the thermophilic methanotroph Methylothermus (91% sequence identity, based on the comparison of 1442 available bases) and the novel halophilic methanotroph Methylohalobius crimeensis (90 %). The other closest neighbors were the moderately thermophilic methanotrophs Methylococcus thermophilus and Methylocaldum szegediense, and the thermotolerant Methylococcus capsulatus Bath (about 85–86% 16S rRNA gene sequence identity). Phylogenetic analysis based on partial pmoA gene sequences confirmed the clustering of M. thermalis with Methylothermus strain HB (90% nucleotide sequence identity and 98% derived amino acid sequence identity) and Methylohalobius crimeensis (83 and 92 %, respectively). In contrast, the partial pmoA gene sequence of M. thermalis exhibited no more than 77% identity to the corresponding gene fragments of Methylocaldum szegediense, Methylocaldum gracile and Methylococcus capsulatus Bath. So far, information on true thermophilic methanotrophs is limited, with only a brief and non-formal taxonomic description of Methylothermus strain HB.
Cell Structure, Metabolism and Life Cycle
The cells grown in liquid culture were mostly represented by coccoids of 0.6–0.8 mm in diameter (Fig. 1a). In contrast, the majority of cells grown on solid medium appeared as short rods, slightly varying in size (0.6–0.8 x 1.0–1.2 mm). The cells were non-motile, and multiplied by normal cell division. The cells showed a typical Gram-negative structure of the cell wall, and the presence of type I intracytoplasmic membrane (ICM) arranged as stacks of vesicular disks (Fig. 1b) that completely filled the cytoplasmic space. Glycogen inclusions and poly-beta-hydroxybutyrate granules usually present in mesophilic methanotrophs were not observed.
Methylothermus thermalis requires methane or methanol as a carbon and energy source to growth. This species can use nitrate, ammonia, urea, tryptophan,lysine, glutamate, formamide and Triss as nitrogen sources, but no growth was obsevered without one of these sources. The cells grew on methane or methanol at 37–67 6C, and optimally at 57–59 6C. Interesting features of cell structure; how it gains energy; what important molecules it produces.
Ecology
biogeochemical significance; This species is a methanotroph and therefore it helps to sequester methane from the atmosphere.
References
Author
Page authored by Anthony Heidt and James Goodrow Jr., student of Prof. Jay Lennon at Michigan State University.
