Escherichia: Difference between revisions
No edit summary |
|||
| Line 1: | Line 1: | ||
{{Biorealm}} | |||
{| | {| | ||
| width="87" height="69" bgcolor="#FFDF95" align="center" | | | width="87" height="69" bgcolor="#FFDF95" align="center" | | ||
Revision as of 14:06, 14 August 2006
A Microbial Biorealm page on the Escherichia
|
NCBI: |
Classification
Higher order taxa:
Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterbacteriaceae
Species:
Escherichia albertii; E. blattae; E. coli; E. fergusonii; E. hermannii; E. senegalensis; E. senegalensis; E. vulneris; E. sp.
Description and Significance
Escherichia coli has become a model organism for studying many of life's essential processes partly due to its rapid growth rate and simple nutritional requirements. Researches have well established information about E. coli's genetics and completed many of its genome sequences - presently, we know more about E. coli than any other living organism. (Microbes in Norwich)
Genome Structure
Several strains of E. coli have been sequenced and studied in detail. E. coli K-12 was the earliest organism to be "suggested as a candidate for whole genome sequencing" (Blattner et al. 1997). It has a single circular chromosome with 4,639,221 base pairs and 4288 protein-coding genes. Of these protein-coding genes, 38% have no attributed function. E. coli K-12's genome, like other E. coli genomes, has a 50.8% G+C content. Genes which coded for proteins account for 87.8% of the genome, stable RNA-encoding genes make up 0.8%, 0.7% is made of noncoding repeats, and about 11% is for regulatory and other functions. Read "The complete genome sequence of Escherichia coli K-12" by Blattner et al. for more in-depth information on the genome of this organism.
Cell Structure and Metabolism
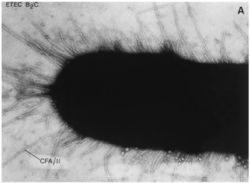
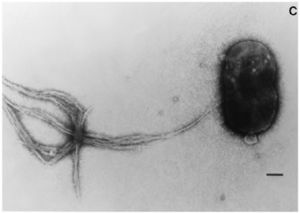
E. coli is a facultative anaerobe that colonizes the lower gut of animals but also survives when released into the envionrment (Blattner et al. 1997). They are rod-shaped bacteria that possess adhesive fimbriae.
E. albertii are nonmotile bacteria that produce acid from the fermentation of D-glucose (with gas), L-arabinose, and D-mannitol and do not ferment sucrose and lactose. They are beta-galactosidase positive and fermented D-arabinose, D-fructose, D-galactose, D-mannose, and ribose; however, they were unable to use many uncommon sugars (Abbott et al. 2003).
Ecology
E. coli typically colonizes an infant's gastrointestinal tract within hours of life and is the predominant facultative anaerobe in the human colonic flora. In this mutual relationship, E. coli generally is confined to the intestinal lumen; however, in a debilitated or immunosuppressed host or when the bacteria is introduced to other tissues, even normal "nonpathogenic" strains of E. coli can cause infection. Other strains of E. coli can cause infection even in a healthy person. This bacterium can be easily grown at 37oC under aerobic conditions and is used in many different types of experiments.
Pathology
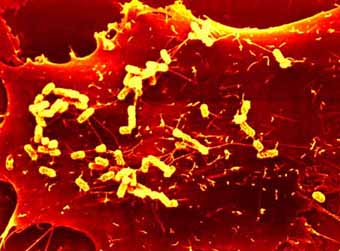
Although other Escherichia are pathogenic, such as E. albertii that is associated with diarrheal disease in Bangladeshi children, E. coli is the most known and studied Escherichia pathogen (Abbott et al. 2003). Nonpathogenic E. coli is generally confined to the intestinal lumen but may cause infection in a debilitated or immunosuppressed host or when the bacteria is introduced to other tissues. Other strains of E. coli, however, can cause infection even in a person who otherwise perfectly healthy. This bacteria can be contracted from undercooked meat or from drinking contaminated water. E. coli infections may be "limited to the mucosal surfaces or can disseminate throughout the body" (Nataro and Kaper 1998). The three general clinical syndromes caused by pathogenic E. coli strains are urinary tract infection, sepsis/meningitis, and enteric/diarrheal disease. The infection strategy of E. coli is to colonize a mucosal site, evade host defenses, multiply, and damage the host. All diarrheagenic strains can colonize the intestinal mucosal surface "despite peristalsis and competition for nutrients by the indigenous flora of the gut" (Nataro and Kaper 1998). Pathogenic and nonpathogenic E. coli bacteria have surface adherence fimbriae, but diarrheagenic E. coli strains have specific fimbrial antigens that adhere to the small bowel mucosa, a site that is normally not colonized. The three general ways in which E. coli may cause diarrhea is by enterotoxin production, invasion, and/or "intimate adherence with membrane signalling" (Nataro and Kaper 1998). For a great article on diarrheagenic E. coli, read "Diarrheagenic Escherichia coli" by Nataro and Kaper.
References
General:
- Microbes in Norwich: Escherichia coli
- Nataro, James P. and James B. Kaper. 1998. "Diarrheagenic Escherichia coli." Clinical Microbiology Reviews, vol. 11, no. 1. American Society for Microbiology. (142-201)
- Blattner, Frederick R., Guy Plunkett III, Craig A. Bloch, Nicole T. Perna, Valerie Burland, Monica Riley, Julio Collado-Vides, Jeremy D. Glasner, Christopher K. Rode, George R. Mayhew, Jason Gregor, Nelson Wayne Davis, Heather A. Kirkpatrick, Michael A. Goeden, Debra J. Rose, Bob Mau, and Ying Shao. 1997. "The complete genome sequence of Escherichia coli K-12." Science, vol. 277. (1453-1462)
Cell Structure and Metabolism:
- Abbott, Sharon L., Jennifer O'Connor, Tom Robin, Barbara L. Zimmer, and J. Michael Janda. 2003. "Biochemical properties of a newly described Escherichia species, Escherichia albertii." Journal of Clinical Microbiology, vol. 41, no. 1. American Society for Microbiology. (4852-4854)