Deinococcus: Difference between revisions
No edit summary |
No edit summary |
||
| (One intermediate revision by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{Curated}} | |||
{{Biorealm Genus}} | {{Biorealm Genus}} | ||
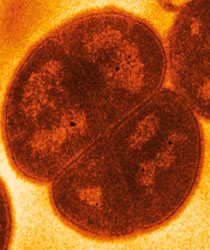
[[Image:deinococcuspic.jpg|frame|right|''D. radiodurans'' from an electron microscope. From [http://www.genomenewsnetwork.org/articles/07_02/deinococcus.shtml Genome News Network.]]] | [[Image:deinococcuspic.jpg|frame|right|''D. radiodurans'' from an electron microscope. From [http://www.genomenewsnetwork.org/articles/07_02/deinococcus.shtml Genome News Network.]]] | ||
| Line 17: | Line 13: | ||
''D. geothermalis, D. grandis, D. indicus, D. murrayi, D. proteolyticus, D. radiodurans, D. radiodurans R1, D. radiophilus, D. radiopugnans, D. sp. D. sp. oral clone CB003, uncultured D. sp. '' | ''D. geothermalis, D. grandis, D. indicus, D. murrayi, D. proteolyticus, D. radiodurans, D. radiodurans R1, D. radiophilus, D. radiopugnans, D. sp. D. sp. oral clone CB003, uncultured D. sp. '' | ||
{| | |||
| height="10" bgcolor="#FFDF95" | | |||
'''NCBI: [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=1298&lvl=3&p=6&p=7&lin=f&keep=1&srchmode=1&unlock Taxonomy] [http://www.ncbi.nlm.nih.gov/genomes/framik.cgi?db=Genome&gi=144 Genome] ''' | |||
|} | |||
==Description and Significance== | ==Description and Significance== | ||
Latest revision as of 21:32, 30 July 2010
A Microbial Biorealm page on the genus Deinococcus
Classification
Higher order taxa:
Bacteria; Deinococcus-Thermus; Deinococci; Deinococcales; Deinococcaceae
Species:
D. geothermalis, D. grandis, D. indicus, D. murrayi, D. proteolyticus, D. radiodurans, D. radiodurans R1, D. radiophilus, D. radiopugnans, D. sp. D. sp. oral clone CB003, uncultured D. sp.
Description and Significance
Deinococcus is a gram-positive bacteria found to form pink or reddish colored colonies. Deinococcus is known for being the most radiation-resistant vegetative cell (R. Murray). In fact, Deinococcus radiodurans can live through blasts of radiation thousands of times greater than the level that would kill a human being, and its Latin name means "strange berry that withstands radiation." To learn how D. radiodurans manage to live through such intense blasts of radiation, go here.
Genome Structure
The genome structure of Deinococcus radiodurans is made up of two chromosomes (2,648,638 and 412,348 base pairs), a megaplasmid (177,466 base pairs), and a small plasmid (45,704 base pairs), creating a genome of 3,284,156 base pairs (TIGR, 2004). The megaplasmid and chromosomes qualities are in part what allows the organism to withstand γ-radiation, desiccation, and oxidizing agents as well as many other DNA-damaging conditions such as starvation. D. radiodurans can also grow at 60Gy/h without any growth rate effects being visible (Y. Lui et al., 2003).
Cell Structure and Metabolism
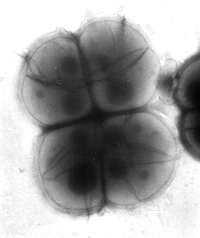
When observed by an electron microscope, the cell wall was discovered to have an unusual thickness of about 50-60 nm, with a dense inner layer of about 14-20 nm. This inner wall creates the septa and sometimes the fenestrated layer. The septum forms an unusual shape, described as a pair of curtains rather than the typical "iris-diaphram". Deinococcus typically forms a tetrad shape, allowing a second division to begin before the first is complete during cell division (R. Murray).
Ecology
The natural habitat of Deinococcus is not yet known because Deinococcus is chemoorganotrophic. It has been isolated from a variety of sites, and needs a complex growth media. It seems that the proteases of Deinococcus may be used for the generation of several useful amino acids and possibly a few sugars as well. Deinococcus strains have been grown from a variety of materials including soil, animal feces, and meat. It is speculated that these aerobic bacteria are likely to live in rich organic habitats, such as feces or intestinal contents (R. Murray).
References
F. J. Franceschi. Max Planck Institute for Molecular Genetics. Large Subunit (50S) Structure.
Genome News Network. 5 July 2002. The World's Toughest Bacterium.
The Institute for Genomic Research: TIGR Microbial Database.2004.
Liu, L., J. Zhou, M. V. Omelchenko, A. S. Beliaev, A. Venkateswaran, J. Stair, L. Wu, D. K. Thompson, D. Xu, I. B. Rogozin, E. K. Gaidamakova, M. Zhai, K. S. Makarova, E. V. Koonin, and M. J. Daly. 2003. Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc Natl Acad Sci U S A. 2003 100: 4191-4196.