Ruminococcus: Difference between revisions
No edit summary |
No edit summary |
||
| (3 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{ | {{Curated}} | ||
{{Biorealm Genus}} | |||
[[Image:g_reaction1.jpg|frame|right|''Ruminococcus albus''. [http://www.genomenewsnetwork.org/articles/04_02/gut_reactions.shtml Photo by Mark Morrison.]]] | [[Image:g_reaction1.jpg|frame|right|''Ruminococcus albus''. [http://www.genomenewsnetwork.org/articles/04_02/gut_reactions.shtml Photo by Mark Morrison.]]] | ||
| Line 14: | Line 13: | ||
''Ruminococcus albus, R. bromii, R. flavefaciens'' | ''Ruminococcus albus, R. bromii, R. flavefaciens'' | ||
{| | |||
| height="10" bgcolor="#FFDF95" | | |||
'''NCBI: [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=1263&lvl=3&lin=f&keep=1&srchmode=1&unlock Taxonomy] [http://www.ncbi.nlm.nih.gov/genomes/framik.cgi?db=Genome&gi=11601 Genome]''' | |||
|} | |||
==Description and Significance== | ==Description and Significance== | ||
Latest revision as of 20:43, 6 August 2010
A Microbial Biorealm page on the genus Ruminococcus
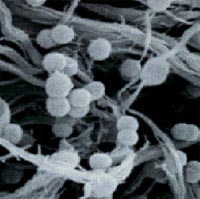
Classification
Higher order taxa:
Bacteria; Firmicutes; Clostridia; Clostridiales; Lachnospiraceae
Species:
Ruminococcus albus, R. bromii, R. flavefaciens
Description and Significance
Ruminococcus is Gram-positive bacteria.
Genome Structure
Work has been started on the Ruminococcus genome structure. In 1995, the genome of Ruminococcus flavefaciens plasmid pBAW301 was sequenced. In 2004, Rincón et. al. discovered a new gene, scaC, in R. flavefaciens. This gene is part of a cluster that codes for ScaA and ScaB, which are components of the cellulosome.
Cell Structure and Metabolism
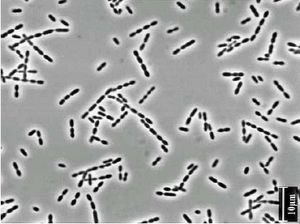
Ruminococcus are non-motile organisms with a coccoid shape.
Ruminococcus are anaerobic bacteria. They obtain nutrients by breaking down cellulose that comes through the digestive system of the host organism. These organisms are also capable of fermenting glucose and xylose.
Ruminococcus do not produce spores.
Ecology
Ruminococcus inhabits the rumen of cattle, sheep, and goats. These organisms allow their hosts to digest cellulose. Ruminococcus' cellulose degredation abilities are currently a major area of study. By understanding how these organisms degrade cellulose, farmers may be able to make advances in animal productivity. In addition, this knowledge could have an environmental impact. It has been suggested that scientists could develop better methods of recycling paper and wood materials.
Simmering et. al. (2002) proposed a new species, Ruminococcus luti, found not in the rumen of cattle, but in human feces. These bacteria are found in chains of up to ten cells. It does not appear to be pathogenic. Cellulolytic Ruminococcus spp. are also detected in many gut habitats including the rumen of goat, sheep and cows and the hindgut of horses, pigs and wild mammals as well as in the human large intestine.
