Thioploca araucae: Difference between revisions
No edit summary |
|||
| (8 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | |||
{{Biorealm Genus}} | {{Biorealm Genus}} | ||
| Line 16: | Line 17: | ||
==Description and significance== | ==Description and significance== | ||
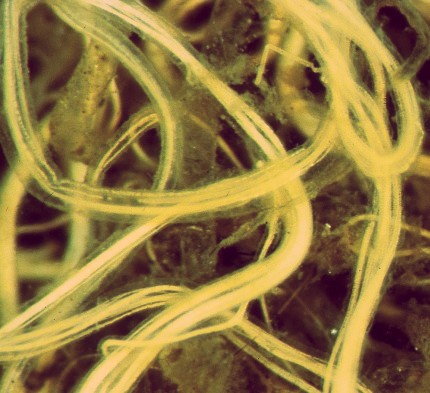
[[image:gia02.jpg]] Sheaths of ''Thioploca araucae'' | |||
''Thioploca araucae'' is a filamentous sulfur-oxidizing bacteria that usually lives in bundles that are surrounded by a sheath. The cells, which are 30-43 um wide and 0.5-1.5 times that in length, are some of the largest bacteria in nature and often have a white or yellow color that can be seen with the naked eye. One filament can contain over a thousand cells, allowing it to reach lengths of up to 7 cm. (9) ''Thioploca'' filaments form thick bacterial mats on the sea floor below the oxygen-minimum zone in upwelling regions of the ocean. Water depths between 40 and 280 meters are common for these areas. (1) The continental shelf off the coast of Chile and southern Peru is one such upwelling region, and is where V.A. Gallardo first discovered ''Thioploca'' in 1977. (5) This marine discovery of ''Thioploca'' was well after the first discoveries of other ''Thioploca'' species in freshwater lakes and ponds. (4) ''Thioploca araucae'' has now been reported in these areas of the oxygen minimum zone underlying the Peru-Chile Subsurface Countercurrent system for a stretch of more than 3000 km and an area exceeding 10,000 km2. This makes it likely that it is the largest community of visible bacteria in the world.(9) | ''Thioploca araucae'' is a filamentous sulfur-oxidizing bacteria that usually lives in bundles that are surrounded by a sheath. The cells, which are 30-43 um wide and 0.5-1.5 times that in length, are some of the largest bacteria in nature and often have a white or yellow color that can be seen with the naked eye. One filament can contain over a thousand cells, allowing it to reach lengths of up to 7 cm. (9) ''Thioploca'' filaments form thick bacterial mats on the sea floor below the oxygen-minimum zone in upwelling regions of the ocean. Water depths between 40 and 280 meters are common for these areas. (1) The continental shelf off the coast of Chile and southern Peru is one such upwelling region, and is where V.A. Gallardo first discovered ''Thioploca'' in 1977. (5) This marine discovery of ''Thioploca'' was well after the first discoveries of other ''Thioploca'' species in freshwater lakes and ponds. (4) ''Thioploca araucae'' has now been reported in these areas of the oxygen minimum zone underlying the Peru-Chile Subsurface Countercurrent system for a stretch of more than 3000 km and an area exceeding 10,000 km2. This makes it likely that it is the largest community of visible bacteria in the world.(9) | ||
Pure cultures of ''Thioploca araucae'' are not in existence, so all samples are on filaments collected from their natural environment. | Pure cultures of ''Thioploca araucae'' are not in existence, so all samples are on filaments collected from their natural environment. | ||
''Thioploca'' sheaths reach a maximum wet weight of 800 grams per meter at a depth of 90 meters. The actual bacterial filaments inside of these sheaths are 10% of the weight, while about 20% of sheaths contain more than one kind of ''Thioploca'' species. (3) '' | ''Thioploca'' sheaths reach a maximum wet weight of 800 grams per meter at a depth of 90 meters. The actual bacterial filaments inside of these sheaths are 10% of the weight, while about 20% of sheaths contain more than one kind of ''Thioploca'' species. (3) ''Thioploca chileae'' is the other major marine species of ''Thioploca''. | ||
''Thioploca araucae'' is important in that it serves as a bridge between the nitrogen and sulfur cycles. It reduces nitrate into elemental sulfur and, subsequently, sulfate. This links the two cylces in the sediment. While ''Thioploca araucae'' is not unique in this respect, it is in that it can store both nitrate and sulfur within its cells. This makes its survival independent of coexisting substrates. (1) | ''Thioploca araucae'' is important in that it serves as a bridge between the nitrogen and sulfur cycles. It reduces nitrate into elemental sulfur and, subsequently, sulfate. This links the two cylces in the sediment. While ''Thioploca araucae'' is not unique in this respect, it is in that it can store both nitrate and sulfur within its cells. This makes its survival independent of coexisting substrates. (1) | ||
| Line 35: | Line 38: | ||
''Thioploca'' cells form filaments that cling to each other and secrete an encompassing sheath of mucous film. The sheath serves as a kind of vertical tunnel through the sediment up to the overlying water, allowing the ''Thioploca'' filaments to glide up and down and thereby commute between their food source and the nitrate they need to metabolize it. Networks of interwoven horizontal and vertical sheaths also form, serving to stabilize the sediment very efficiently. ''Thioploca araucae'' and its sheaths | ''Thioploca'' cells form filaments that cling to each other and secrete an encompassing sheath of mucous film. The sheath serves as a kind of vertical tunnel through the sediment up to the overlying water, allowing the ''Thioploca'' filaments to glide up and down and thereby commute between their food source and the nitrate they need to metabolize it. Networks of interwoven horizontal and vertical sheaths also form, serving to stabilize the sediment very efficiently. ''Thioploca araucae'' and its sheaths are often mixed in with other bacteria. Among these are ''Thioploca chileae'', the other marine species of ''Thioploca'', and ''Beggiatoa''. In 85% of the sheaths that have mixed populations, ''Thioploca araucae'' is more widespread. In fact, the ''Thioploca araucae'' population is larger in both biomass and number than the ''Thioploca chileae'' population (3). | ||
By using its sheaths as a transport system, ''Thioploca araucae'' is able to put nitrate deep into the seafloor. This eliminates competition from other sulfide oxidizing bacteria, which cannot store an electron acceptor for long periods of time. Those bacteria need simultaneous access to both electron acceptors and electron donors. ''Thioploca'' can commute up and down, gaining nitrate at higher depths and oxidizing sulfide at lower levels. Elemental sulfur is stored as an energy reserve. In this way, ''Thioploca araucae'' is able to overcome the issue of substrate availability.(9) | By using its sheaths as a transport system, ''Thioploca araucae'' is able to put nitrate deep into the seafloor. This eliminates competition from other sulfide oxidizing bacteria, which cannot store an electron acceptor for long periods of time. Those bacteria need simultaneous access to both electron acceptors and electron donors. ''Thioploca'' can commute up and down, gaining nitrate at higher depths and oxidizing sulfide at lower levels. Elemental sulfur is stored as an energy reserve. In this way, ''Thioploca araucae'' is able to overcome the issue of substrate availability.(9) | ||
| Line 60: | Line 63: | ||
Another process that can occur in bacteria that have a nutritional imbalance is the accumulation of phosphate. ''Thioploca'' shows efficient phosphate uptake and storage, and also contains PolyP storage granules. This has lead to hypotheses that many phosphorite deposits on the sea floor can be credited to ''Thioploca'' and the other large sulfur-oxidizing bacteria. Heide and Horst Shulz look into this and suggest a mechanism for the release of phosphate into the anoxic sediment by these bacteria. Through the staining of cells with toluidine blue, all of these sulfur bacteria species were shown to contain large amounts of intracellular polyphosphates. The bacteria also showed electron-dense inclusions, a typical appearance for polyphosphates. New phosphorite formation has also been reported in areas that contain large populations of ''Thioploca'', ''Beggiatoa'', and/or ''Thiomargarita''. Working mainly with ''Thiomargarita namibiensis'', they were able to show a steady rate of phosphate release under anoxic conditions. (12) | Another process that can occur in bacteria that have a nutritional imbalance is the accumulation of phosphate. ''Thioploca'' shows efficient phosphate uptake and storage, and also contains PolyP storage granules. This has lead to hypotheses that many phosphorite deposits on the sea floor can be credited to ''Thioploca'' and the other large sulfur-oxidizing bacteria. Heide and Horst Shulz look into this and suggest a mechanism for the release of phosphate into the anoxic sediment by these bacteria. Through the staining of cells with toluidine blue, all of these sulfur bacteria species were shown to contain large amounts of intracellular polyphosphates. The bacteria also showed electron-dense inclusions, a typical appearance for polyphosphates. New phosphorite formation has also been reported in areas that contain large populations of ''Thioploca'', ''Beggiatoa'', and/or ''Thiomargarita''. Working mainly with ''Thiomargarita namibiensis'', they were able to show a steady rate of phosphate release under anoxic conditions. (12) | ||
Marc Mussman and his colleagues are delving into ways to get a better understanding of the genomes of ''Thioploca araucae'' and other similar sulfur-oxidizing bacteria. They propose using a combination of optical mapping, WGA, and pyrosequencing as the best way to go about analysis of individual, uncultured bacteria. However, the incomplete sequence assemblies still prevent the accurate determination of the genome size and an in-depth analysis of the ''Thioploca'', ''Beggiatoa'', and ''Thiomargarita'' genomes. Often, the contribution of non-target DNA cannot be completely ruled out in environmental WGA projects because they have difficulty in differentiating between lone cells that are not within sheaths or other attachment complexes(7) | Marc Mussman and his colleagues are delving into ways to get a better understanding of the genomes of ''Thioploca araucae'' and other similar sulfur-oxidizing bacteria. They propose using a combination of optical mapping, WGA, and pyrosequencing as the best way to go about analysis of individual, uncultured bacteria. However, the incomplete sequence assemblies still prevent the accurate determination of the genome size and an in-depth analysis of the ''Thioploca'', ''Beggiatoa'', and ''Thiomargarita'' genomes. Often, the contribution of non-target DNA cannot be completely ruled out in environmental WGA projects because they have difficulty in differentiating between lone cells that are not within sheaths or other attachment complexes.(7) | ||
==References== | ==References== | ||
| Line 90: | Line 93: | ||
13. NCBI: Thioploca araucae. Accessed August 28, 2007. <http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=439499> | 13. NCBI: Thioploca araucae. Accessed August 28, 2007. <http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=439499> | ||
Edited by student of [mailto:ralarsen@ucsd.edu Rachel Larsen] | Edited by Kyle Backer, student of [mailto:ralarsen@ucsd.edu Rachel Larsen] | ||
Edited by KLB | |||
Latest revision as of 03:37, 20 August 2010
A Microbial Biorealm page on the genus Thioploca araucae
Classification
Cellular Organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Thiotrichales; Thiotrichaceae; Thioploca (13)
Species
|
NCBI: Taxonomy |
Thioploca araucae
Description and significance
Thioploca araucae is a filamentous sulfur-oxidizing bacteria that usually lives in bundles that are surrounded by a sheath. The cells, which are 30-43 um wide and 0.5-1.5 times that in length, are some of the largest bacteria in nature and often have a white or yellow color that can be seen with the naked eye. One filament can contain over a thousand cells, allowing it to reach lengths of up to 7 cm. (9) Thioploca filaments form thick bacterial mats on the sea floor below the oxygen-minimum zone in upwelling regions of the ocean. Water depths between 40 and 280 meters are common for these areas. (1) The continental shelf off the coast of Chile and southern Peru is one such upwelling region, and is where V.A. Gallardo first discovered Thioploca in 1977. (5) This marine discovery of Thioploca was well after the first discoveries of other Thioploca species in freshwater lakes and ponds. (4) Thioploca araucae has now been reported in these areas of the oxygen minimum zone underlying the Peru-Chile Subsurface Countercurrent system for a stretch of more than 3000 km and an area exceeding 10,000 km2. This makes it likely that it is the largest community of visible bacteria in the world.(9) Pure cultures of Thioploca araucae are not in existence, so all samples are on filaments collected from their natural environment. Thioploca sheaths reach a maximum wet weight of 800 grams per meter at a depth of 90 meters. The actual bacterial filaments inside of these sheaths are 10% of the weight, while about 20% of sheaths contain more than one kind of Thioploca species. (3) Thioploca chileae is the other major marine species of Thioploca. Thioploca araucae is important in that it serves as a bridge between the nitrogen and sulfur cycles. It reduces nitrate into elemental sulfur and, subsequently, sulfate. This links the two cylces in the sediment. While Thioploca araucae is not unique in this respect, it is in that it can store both nitrate and sulfur within its cells. This makes its survival independent of coexisting substrates. (1)
Genome structure
The genome is currently being sequenced as part of the Moore Foundation Microbial Genome Sequencing project. A sample collected by Victor Gallardo on the continental shelf off of Concepcion, Chile is being studied by the J Craig Venter Institute. (13) Data from 16S rRNA analysis suggests that Beggiatoa is Thioploca's closest phylogenetic relative. Thioploca and Beggiatoa form a phylogenetic lineage within the gamma-subdivision of the proteobacteria, showing similar genetic diversity that other groups of sulfur oxidizers have. Often differentiated simply by size, fluorescent in situ hybridization with Thioploca-specific oligonucleotide probes can used to separate the sequences of Thioploca araucae and Thioploca chileae. (10)
Cell structure and metabolism
Thioploca araucae contain light-refracting sulfur globules in their cytoplasm that surround a large vacuole. This vacuole is used to concentrate nitrate at levels up to 500 mM and can occupy greater than 80% of the cell's volume.(1) Thus, Thioploca araucae can store both sulfur and nitrogen products within its cytoplasm. Gliding filaments serve to trasport this nitrate 5-10 cm down into the sediment, where it will be reduced.(3) A sequence of reactions is then carried out in order to reduce nitrate and oxidize sulfur. For denitrification, hydrogen sulfide is first oxidized using the nitrate, resulting in the production of elemental sulfur that is stored in the cytoplasm in the form of the sulfur globules. This reaction is thought to be: 2NO3- + 5HS. + 7H+ --> N2 + 5S + 6H2O. The elemental sulfur is then oxidized, resulting in sulfate: 6NO3- + 5S + 2H2O --> 3N2 + 5SO4(2-) + 4H+. (1) Nitrate ammonification is another series of reactions that can occur to break down the nitrate, resulting in ammonia as a product instead of nitrogen gas. The first reaction involves: 4H2S + NO3- + 2H+ --> 4S + NH4+ + 3H2O. This is followed by: 7H2O + 3NO3- + 4S --> 3NH4+ + 4SO4(2-) + 2H+. The sulfur compound hydrogen sulfide serves as a source of electrons for bacterial chemosynthesis, while the oxidation of sulfur serves as a source of energy instead of light. The result of this is a linking of the nitrogen and sulfur cycles.
Ecology
Thioploca cells form filaments that cling to each other and secrete an encompassing sheath of mucous film. The sheath serves as a kind of vertical tunnel through the sediment up to the overlying water, allowing the Thioploca filaments to glide up and down and thereby commute between their food source and the nitrate they need to metabolize it. Networks of interwoven horizontal and vertical sheaths also form, serving to stabilize the sediment very efficiently. Thioploca araucae and its sheaths are often mixed in with other bacteria. Among these are Thioploca chileae, the other marine species of Thioploca, and Beggiatoa. In 85% of the sheaths that have mixed populations, Thioploca araucae is more widespread. In fact, the Thioploca araucae population is larger in both biomass and number than the Thioploca chileae population (3).
By using its sheaths as a transport system, Thioploca araucae is able to put nitrate deep into the seafloor. This eliminates competition from other sulfide oxidizing bacteria, which cannot store an electron acceptor for long periods of time. Those bacteria need simultaneous access to both electron acceptors and electron donors. Thioploca can commute up and down, gaining nitrate at higher depths and oxidizing sulfide at lower levels. Elemental sulfur is stored as an energy reserve. In this way, Thioploca araucae is able to overcome the issue of substrate availability.(9)
It is estimated that close to one-quarter of global marine denitrification is done in the upwelling waters off the Pacific coast of South America (2). Thioploca araucae, along with other sulfur oxidizers such as Beggiatoa, plays a major role in this. Also, close physical associations between Thioploca and filamentous gamma-proteobacteria have been observed in organic-rich marine sediments, where Desulfonema filaments are epibionts on the larger sulfur-oxidizing filaments like Thioploca araucae. (8) Inhabited sheaths of Thioploca are often heavily covered with the smaller Desulfonema.
As a result of Thioploca araucae's large range, size, efficiency of nitrate intake, and ability to unite the nitrogen, sulfur, and carbon cycles, it is assumed that it plays a large role in the biogeochemistry of marine upwelling areas. This, however, remains to be proven. The fact that Thioploca is turning up in more places all the time, including the Arabian Sea, off the Namibian coast, and around geothermal vents in the Mediterranean Sea, lends credence to the thought that it plays a significant role in shaping the composition of anoxic sediments. (9)
Thioploca araucae has also been found in the mouths and guts of marine animals such as shrimp. This, coupled with the large biomass of the Thioploca, suggests a trophic relationship between animals and Thioploca. (11)
Pathology
There is no known pathogenic function for Thioploca araucae.
Application to Biotechnology
Nothing specific is noted as an application to biotechnology, but continued study of the mechanisms involved in linking the carbon, nitrogen, and sulfur cycles could eventually give rise to advances in other fields.
Current Research
Most current research involves studying what conditions are favored by Thioploca araucae, its actual method of metabolizing sulfur and nitrogen, and the ongoing quest for its genome. Many comparative studies are also being undertaken to determine what differences and similarities there are between the closely related genera of Thioploca, Beggiatoa, and Thiomargarita.
Tobin Strohm's research involves studying at what points will Thioploca utilize denitrification and at which points will it utilize nitrate ammonification. Under conditions of limited nitrate availability, Thioploca and other sulfur-oxidizing bacteria seem to prefer nitrate ammonification. They do so at a rate of two to one, with 15.6 g of ammonia produced per each reduced nitrate as opposed to 7.6 g of nitrogen produced per each reduced nitrate. This happens despite the fact that denitrification should provide the Thioploca with more energy if free energy changes of the overall redox reactions (delta g) are taken into account. Energy yields of nitrate reduction end up being much lower than one would expect. Lithoautotrophic sulfide oxidation such as that undergone by Thioploca araucae tends to prefer nitrate ammonification to denitrification. (6)
The genera Thioploca, Beggiatoa, and Thiomargarita are among the largest prokaryotes known, and they usually contain a vacuole that can account for up to 90% of the cell volume. On the seafloor these large sulfur-oxidizing bacteria fulfill an important ecological function by preventing the release of toxic hydrogen sulfide from the sediment into the water column. Also, when experiencing anoxia these large vacuolated Beggiatoa, Thioploca, and Thiomargarita respire nitrate, which they concentrate up to 10,000-fold (~500 mM) within their intracellular vacuoles.
Another process that can occur in bacteria that have a nutritional imbalance is the accumulation of phosphate. Thioploca shows efficient phosphate uptake and storage, and also contains PolyP storage granules. This has lead to hypotheses that many phosphorite deposits on the sea floor can be credited to Thioploca and the other large sulfur-oxidizing bacteria. Heide and Horst Shulz look into this and suggest a mechanism for the release of phosphate into the anoxic sediment by these bacteria. Through the staining of cells with toluidine blue, all of these sulfur bacteria species were shown to contain large amounts of intracellular polyphosphates. The bacteria also showed electron-dense inclusions, a typical appearance for polyphosphates. New phosphorite formation has also been reported in areas that contain large populations of Thioploca, Beggiatoa, and/or Thiomargarita. Working mainly with Thiomargarita namibiensis, they were able to show a steady rate of phosphate release under anoxic conditions. (12)
Marc Mussman and his colleagues are delving into ways to get a better understanding of the genomes of Thioploca araucae and other similar sulfur-oxidizing bacteria. They propose using a combination of optical mapping, WGA, and pyrosequencing as the best way to go about analysis of individual, uncultured bacteria. However, the incomplete sequence assemblies still prevent the accurate determination of the genome size and an in-depth analysis of the Thioploca, Beggiatoa, and Thiomargarita genomes. Often, the contribution of non-target DNA cannot be completely ruled out in environmental WGA projects because they have difficulty in differentiating between lone cells that are not within sheaths or other attachment complexes.(7)
References
1. H. Fossing, V.A. Gallardo, et. al. "Concentration and transport of nitrate by the mat-forming sulphur bacterium Thioploca". Nature. 1995. Volume 374. p.713-715.
2. Codispoti, L.A. et al. "High Nitrite Levels off Northern Peru: A Signal of Instability in the Marine Denitrification Rate". Science. 1986. Volume 233. p.1200-1202.
3. Schulz, H., Jorgensen, B., Fossing, H., and Ramsing, N. "Community Structure of Filamentous, Sheath-Building Sulfur Bacteria, Thioploca spp., off the Coast of Chile". Applied and Environmental Microbiology. 1996. Volume 62. p. 1855-1862.
4. Lauterborn, R. "Eine neue Gattung der Schwefelbakterien (Thioploca Schmidlei nov. gen. nov. spec.)". Ber. Dtsch. Bot. Ges. 1907. Volume 25. p.238–242.
5. Gallardo, V. A. "Large benthic microbial communities in sulphide biota under Peru-Chile Subsurface Countercurrent". Nature (London). 1997. Volume 268. p.331–332.
6. Strohm, T., Griffin, B., Zumft, W., and Schink, B. "Growth Yields in Bacterial Denitrification and Nitrate Ammonification". Applied and Environmental Microbiology. 2007. Volume 73(5). p. 1420-1424.
7. Mußmann, M., Hu, F., Richter, M., et al. "Insights into the Genome of Large Sulfur Bacteria Revealed by Analysis of Single Filaments". PLoS Biology. 2007. Volume 5(9). e230.
8. Macalady, J., et al. "Dominant Microbial Populations in Limestone-Corroding Stream Biofilms, Frasassi Cave System, Italy". Applied and Environmental Microbiology. 2006. Volume 72(8). p. 5596-5609.
9. Jørgensen, B. and Gallardo, V. "Thioploca spp. filamentous sulfur bacteria with nitrate vacuoles". FEMS Microbiology Ecology. 1999. Volume 28(4), p. 301-313.
10. Teske, A., Ramsing, N. B., Kuver J., and Fossing, H. "Phylogeny of Thioploca and related filamentous sulfide-oxidizing bacteria". Syst. Appl. Microbiology. 1996. Volume 18. p. 517-526.
11. Larking, J., and Strohl, W. "Beggiatoa, Thiothrix, and Thioploca". Ann. Rev. Microbiology. 1983. Volume 37. p. 361-362.
12. Schulz, Heide, and Schulz, Horst "Large Sulfur Bacteria and the Formation of Phosphorite". Science. 2005. Volume 307. p. 416-418.
13. NCBI: Thioploca araucae. Accessed August 28, 2007. <http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=439499>
Edited by Kyle Backer, student of Rachel Larsen
Edited by KLB