Saccharomyces cerevisiae NEU2011: Difference between revisions
No edit summary |
No edit summary |
||
| Line 92: | Line 92: | ||
'''Saccharomyces cerevisiae and Development of Efficient Biofuels:''' | '''Saccharomyces cerevisiae and Development of Efficient Biofuels:''' | ||
[[Image:Biofuel.jpg|thumb|750px|alt=Alt| ''Figure 5: Co-fermentation of Xylose and Cellubiose in Saccharomyces cerevisiae for efficient ethanol production from plant cellulose for biofuels.'' [http://0-www.pnas.org.ilsprod.lib.neu.edu/content/108/2/504.full]]] | |||
The efficient utilization of cellulose from plant material for biofuel production has been a popular subject in modern research. Cellulose contains many different hexoses (such as glucose) and pentoses (such as xylose), that can usually be fermented independently by various organisms, however the co-fermentation of the two has not been an efficient process thus far. In 2010, Suk-Jin Ha et al., worked to overcome this inefficiency by engineering a Saccharomyces cerevisiae to ferment both cellobiose (a glucose dimer) and xylose concurrently. Normally, S. cerevisiae that is used in ethanol biofuel production can efficiently ferment glucose as a carbon source, but not xylose since it lacks the necessary metabolic pathways The genes XYL1 and XYL2 encode the proteins xylose deductase (XR) and xylitol dehydrogenase (XDH) respectively, and the introduction of these genes into S. cerevisiae from Pichia stipitis can create a xylose hydrolysis pathway in the cell. However, now that the organism now had the ability to ferment both glucose and xylose, S. cerevisiae will still ferment glucose preferentially. In addition, once the glucose is depleted and a chance for xylose fermentation could occur, the ethanol levels are usually already so high that further fermentation is suppressed. To bypass this, Suk-Jin Ha et al also engineered a pathway into S. cerevisiae that would hydrolyze cellubiose instead of the glucose monomer. This was achieved by inserting the genes for a high affinity cellodextrin transporter, which is used for bringing cellubiose into the cell, and β-glucosidase, which hydrolyzes the cellubiose into glucose. Figure 5 displays the cofermentation of xylose and cellubiose in the cell. This engineered S. cerevisiae was shown to yield much higher levels of ethanol that S. cerevisiae that can only ferment glucose. This overall process makes the fermentation of plant cellulose into ethanol a much more efficient and useful method of biofuel production than it had ever been considered before [http://0-www.pnas.org.ilsprod.lib.neu.edu/content/108/2/504.full 31]. | The efficient utilization of cellulose from plant material for biofuel production has been a popular subject in modern research. Cellulose contains many different hexoses (such as glucose) and pentoses (such as xylose), that can usually be fermented independently by various organisms, however the co-fermentation of the two has not been an efficient process thus far. In 2010, Suk-Jin Ha et al., worked to overcome this inefficiency by engineering a Saccharomyces cerevisiae to ferment both cellobiose (a glucose dimer) and xylose concurrently. Normally, S. cerevisiae that is used in ethanol biofuel production can efficiently ferment glucose as a carbon source, but not xylose since it lacks the necessary metabolic pathways The genes XYL1 and XYL2 encode the proteins xylose deductase (XR) and xylitol dehydrogenase (XDH) respectively, and the introduction of these genes into S. cerevisiae from Pichia stipitis can create a xylose hydrolysis pathway in the cell. However, now that the organism now had the ability to ferment both glucose and xylose, S. cerevisiae will still ferment glucose preferentially. In addition, once the glucose is depleted and a chance for xylose fermentation could occur, the ethanol levels are usually already so high that further fermentation is suppressed. To bypass this, Suk-Jin Ha et al also engineered a pathway into S. cerevisiae that would hydrolyze cellubiose instead of the glucose monomer. This was achieved by inserting the genes for a high affinity cellodextrin transporter, which is used for bringing cellubiose into the cell, and β-glucosidase, which hydrolyzes the cellubiose into glucose. Figure 5 displays the cofermentation of xylose and cellubiose in the cell. This engineered S. cerevisiae was shown to yield much higher levels of ethanol that S. cerevisiae that can only ferment glucose. This overall process makes the fermentation of plant cellulose into ethanol a much more efficient and useful method of biofuel production than it had ever been considered before [http://0-www.pnas.org.ilsprod.lib.neu.edu/content/108/2/504.full 31]. | ||
Revision as of 00:50, 11 April 2011
Template:Biorealm Genus Saccharomyces cerevisiae
Classification
Higher order taxa
Domain: Eukarya
Kingdom: Fungi
Subkingdom: Dikarya
Phylum: Ascomycota
Subphylum: Saccharomycotina
Class: Saccharomycetes
Order: Saccharomycetales
Family: Saccharomycetaceae
Genus: Saccharomyces
Species: Cerevisiae
Species
| Taxonomy of Saccharomyces cerevisiae |
Genus species
Description and significance
Saccharomyces cerevisiae is a single-celled Eukaryotic budding yeast belonging to the Ascomycetes, a highly diverse group of fungi. The average cell measures approximately 8 micrometers in diameter, and they typically display a round to ovoid morphology [4]. S. cerevisiae is an important model organism as it was the first Eukaryote to have its genome fully sequenced. (2)
The most important commercial use for S. cerevisiae would be in food production. In the presence of oxygen, yeast cells can reproduce and CO2 by ways of the Citric Acid Cycle. When oxygen is not abundant, S. cerevisiae can also metabolize and reproduce, resulting in CO2 and alcohol by ways of fermentative glycolysis [4]. Used as the primary fermenter in brewing and as a leavening agent in baking, S. cerevisiae ferments sugars into carbon dioxide and alcohol during the brewing and baking processes. Because of these functions, S. cerevisiae is commonly referred to as “Baker’s yeast” or “Brewer’s yeast”. Additionally, the genus name “Saccharomyces” is derived from Latinized Greek and translates to “Sugar Fungus” [2]. The utilization of S. cerevisiae can be dated back to 3150 BC, where strains have been isolated from jars of King Scorpion I 5.
In addition to brewing and breadmaking, Saccharomyces cerevisiae has many pharmaceutical functions. The genome can be engineered to overproduce certain metabolites and commercially important compounds for the use in pharmaceutical agents. Examples of drugs that S. cerevisiae can express compounds for include naringenin, coumarate, artemisinin, taxol, amorphadiene and vitamin C 8.
In nature, S. cerevisiae exists in dense populations where in areas where damaged fruits are located, such as vineyards and fruit farms. S. cerevisiae has additionally been isolated from habitats such as soil, in rivers, and even in the human immune system. It can primarily be found in sugar rich environments, which are ideal for fermentation and reproduction. 3
S. cerevisiae can exist in several different strains. S288c was the first “modern” form of this yeast to be developed in a laboratory setting by Dr. Robert Mortimer the 1950s. As opposed to previously studied strains of yeast, this strain was heterothallic, meaning that the species has different sexes that occur in different individuals, and was therefore adopted as a useful model organism for recombinant research. 3 The S288c strain was later used to isolate other strains of S. cereviae in nature. In order to develop this particular strain, Dr. Mortier performed cross breeding of another heterothallic strain called EM93 that was isolated from rotten figs by Dr. Emil Mrak 3.
Genome structure
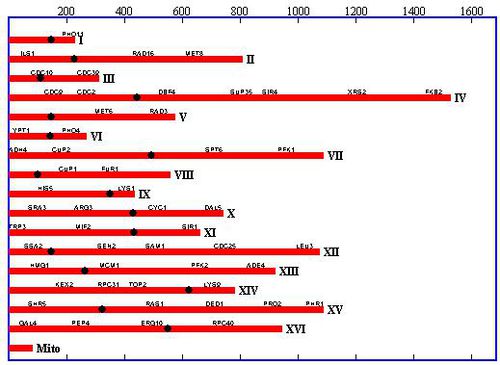
In 1996, Saccromyces cerevisiae was the first Eukaryote to have its genome completely sequenced by a team of over 600 scientists worldwide 5. As of that time, it was the largest genome to be sequenced to date. Because of this, the S. cerevisiae genome has been studied extensively and serves as a model organism for other genomic research. The genome consists of 16 chromosomes (8 pairs) in its haploid vegetative state (as displayed in figure 2). In nature, two of these haploid cells will fuse to form a diploid cell with 32 chromosomes (16 pairs). The haploid stage can exist in two forms, α and a, and two cells of the opposite form will fuse to form the diploid cell. S. cerevisiae haploid cells have the ability to switch between the two haploid mating types. The MAT, aka mating type, operon on the S. cerevisiae genome determines which mating type will be expressed. This mating process can produce great genetic variability from generation to generation [4].
When the genome was originally sequenced, it was assumed that all open reading frames longer than 100 codons would code for a protein. In the 15 years to follow, that hypothesis has been continually tested and the genome itself has been updated to contain 522 new ORFs that the originally sequenced genome did not account for 7. On February 2, 2011, an updated release of the S. cerevisiae genome was released 6. This data shows that 70% of the 12,157 kilobase S. cerevisiae genome encodes a protein. This amounts to 4,902 verified open reading frames. In addition to this, 27 genes are known to encode for rRNA, 6 encode for snRNA, and 299 encode for tRNA. This RNA encoding genes are scattered throughout the 16 chromosomes. The updated version of the genome, which can be found at yeastgenome.org, was sequenced using modern Illumina HiSeq, which outdates the Sanger sequencing methods used in 1996. The complete genomes of all major lab strains of S. cerevisiae are expected in the near future using the modern methods.
Cell structure, Metabolism & Life Cycle
Saccharomyces cerevisiae is a eukaryotic yeast that consists of a single cell, which is either round or ovoid and approximately 4-10 micrometers. The diploid variant is about 5-6 micrometers and is ovoid-shaped. When the diploid cells lack the necessary amounts of nitrogen, psuedohyphal growth may occur under direction of theheterotrimeric G protein alpha hamolog, GPA2 16.
S. cerevisiae has a cell wall composed of two layers, which are made up of mostly polysaccharides, but it is also contain lipids and proteins 15. The inner layer provides mechanical strength to the wall and supplies attachment sites for the proteins that form the outer layer. The inner layer is very strong because it is composed of β1,3-glucan and chitin. Exterior to the inner layer is the outer cell wall which provides osmotic and physical protection to the cell. The outer layer is composed of glycosylated mannoproteins that originate from the surface of the cell. These mannoproteins limit the ability of foreign enzymes to access the inner layer of the cell. The β1,3-glucan complexes have extensive branching, which further explains the strength of the cell wall. The cell wall is about 300 nanometers thick, and is smooth with the exception of bud scars, which forms on the mother cell after a daughter cell buds off. Other essential organelles to the cell include the nucleus, vacuoles, endoplasmic reticulum, and the mitochondria 14.
An integral characteristic of S. cerevisiae is that it is able to utilize two different metabolic pathways: aerobic respiration and anaerobic fermentation. S. cerevisiae is considered a heterotrophic organism because it breaks down carbohydrates to use as energy 17. S. cerevisiae performs respiration or fermentation depending on the presence or absence of oxygen (4). When undergoing respiration, S. cerevisiae completely reduces pyruvate to carbon dioxide, utilizing oxygen as the final electron accpetor. This provides the organism with the largest amount of ATP. When S. cerevisiae undergoes anaerobic respiration, glucose is fermented to produce ethanol 17. Fermentation will only occur under anoxic conditions because respiration yields much more energy per glucose than does fermentation (4) Fermentation produces carbon dioxide and ethanol, which explain the carbonation and alcohol found in beer 17.
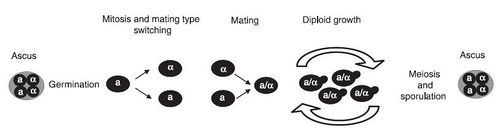
The life cycle of S. cerevisiae has not yet been studied in nature, however it has been extensively studied in a laboratory setting. The life cycle begins as a unicellular diploid that reproduces through budding. Under times of environmental stress, meiosis occurs and results in four haploid spores. These spores are called ascus. The haploid spores can be one of two “mating types”, a or α, which is determined by the MAT genetic locus. The combination of two asci of different mating types will result in a diploid vegetative cell. S. cerevisiae also has an HO gene, which allows asci to switch mating types. Strains with this gene are referred to as homothallic (figure 3). As a result of this, a mother cell can switch mating types and fuse with the daughter cell after division. This inbreeding can result in a loss of heterozygosity 3.
Ecology

Due to its extensive use in human applications such as brewing, baking, distilling, and winemaking, it was once argued that there were no longer any wild strains of S. cerevisiae in nature 9. This is because the extent of what was known about the natural habitat of S. cerevisiae seemed to be limited to fruit fermentation in vineyards. If any wild strains did exist outside of these domestic environments, it was thought that they had evolved from the domestic strain, instead of the other way around 9. However, these presumptions changed as more research was conducted. Today, strains of Saccharomyces yeast have been found in a broad range of habitats such as in the soil of oak trees in New England 10, in the Danube river of Europe 11, and amongst trees and fruits in Asia 3.
Although found in a variety of habitats, the most is known about the role of S. cerevisiae in the vineyard (figure 4). In this environment they thrive on damaged grapes where they can carry out fermentation of the fruit sugars into ethanol and CO2. Fermentation is a very rapid and efficient process in Saccharomyces and has been shown to produce as much ATP per second as aerobic metabolism in other species 13. Ethanol fermentation is not unique to S. cerevisiae, however its ability ferment at such a rapid pace in the harsh grape environment is. Grape juice is very acidic with a pH between 2.5 and 3.8 caysed by tartrate, lactate, and malate acids. There is also a very high osmotic pressure because of the high sugar content in the environment. In addition, grapes grow best in environments with a range of temperatures and humidity levels so S. cerevisiae must be tolerant these fluctuations. Nitrogen and oxygen are also very scarce. The combination of these factors (high acidity, high osmotic pressure, anaerobic, and temperate variation) make it hard for any other organism to compete with S. cerevisiae in this environment once fermentation has begun. In addition, S. cerevisiae has a much higher tolerance to ethanol concentrations in the environment so their fermentation can actually kill off other species 3.
Yeast also has a competitive advantage in vineyards because it has been shown to produce a toxin that is lethal to other bacteria and more sensitive yeast species. Because of this quality, S. cerevisiae is classified in a group called “Killer yeasts”4. This “killer” quality is most advantageous in a setting that is very dense with other organisms, however they have been shown to ferment and grow more slowly once in isolation. The toxins created by these yeast cells resemble viruses that damage the plasma membrane of neighboring cells 12.
Since S. cerevisiae has such a tight relationship with human industry, it is thought that its genetics and geographic distribution have been greatly altered from the wildtype over time. However, S. cerevisiae has a very closely related sister species, S. paradoxus, which has little to no association with human influences. These two species are estimated to have diverged from a common ancestor 5-10 million years ago and are nearly phenotypically identical. Therefore, scientists often look to this species as a model ecological organism while looking to S. cerevisiae as a model genetic organism 12.
Pathogenicity
Although Saccharomyces cervisiae is considered to be nonpathogenic 18, it has been known to cause food spoilage. S. cerevisiae has been found in isolated samples of spoiled fruit juices, sports drinks as well as dairy products 19. When a fungi is genotypically and phenotypically diverse enough to adapt to a certain environment, such as the human body, it is a concern that the fungi could potentially become pathogenic 20. Despite the rarity of infection, since 1980 there have been reports of infections attributed to S. cerevisiae 21. It is now known that S. cerevisiae infects human patients on both sterile and non-sterile body sites, suggesting that at least some isolates do have the potential to invade and proliferate like a pathogen 20. Invasive infections were reported in patients that were severely debilitated and immunodeficient. Clinical isolates in competent outbred mice, S. cerevisiae have the ability to proliferate and persist. Further, S. cerevisiae have the ability to grow at high temperatures (42°C), an indication for virulence 21.
Current Research
Saccharomyces cerevisiae and Antiviral Drug Screening:
In 2009, Schneider and colleagues performed a metabolite profiling study on Saccharomyces cerevisiae. Their study used S. cerevisiae to prioritize host targets for antiviral drug screening. The target hosts tested were cellular proteins Pat1p, Lsm1p, and Dhh1p. These three proteins were tested on reference strains BY4742 Matα his3∆1 leu2∆0 lys2∆0 ura3∆0 and deletion strains pat1∆, lsm1∆, and dhh1∆. Using gas chromatography/mass spectrometry (GC/MS) analysis, 41 out of 47 polar metabolites were identified. Using partial least squares to latent structures discriminate analysis (PLS-DA) researchers were able to identify metabolites with significant variations between different strains. Deletion strain pat1∆ was identified as having the least number of differences and growth rates, and specific production rates of ethanol and glycerol most similar to the reference strain. Based on these results, Schneider and his colleagues hypothesized that the human analog of yeast pat1Δ is the best drug target candidate 22.
Saccharomyces cerevisiae and Lactating Dairy Cows:
When lactating dairy cows are subjected to heat stress, particularly above a temperature humidity index (THI) of 72, they have decreased intake of impact dry matter (DM) diet and overall milk production. The normal temperature range of a dairy cow is 5 to 25°C. One study showed that when dairy cows were fed the mold fungi Aspergillus oryzae during the summer, there was reduced temperature measured rectally as well as an increase in milk production.
Bruno and colleagues repeated the experiment on 725 early lactation dairy cows in San Joaquin Valley, California using a culture of Saccharomyces cerevisae. Cows were fed either a DM diet with or without the added yeast culture. The diet of 30g of a culture of S. cerevisae was given twice per day and intake, temperature and humidity were recorded. For the first 20 days, the cows were milked four times per day and during the 120 day study, the cows were milked two times per day. During June to September, the cows were under heat stress with THI above 72.
The study showed that when subjected to heat stress, dairy cows given the S. cerevisae treatment had a lower rectal temperature by 0.2°C compared to the control group. The intake of DM did not differ significantly between the control group and the S. cerevisae group. However, the S. cerevisae cows produced 1.2kg of milk per day more than control milk cows consistently during the 120 day study period. This increase could be due to S. cerevisae improving rumen pH and digestibility of the DM and increasing energy for milk synthesis. Bruno and colleagues concluded that even the small decrease in rectal temperature suggests an improved tolerance to heat stress and an increase in milk production suggests S. cerevisae is an effective nutritional supplement when daily cows are subject to heat stress during warm summer months 23.
Saccharomyces cerevisiae and Development of Efficient Biofuels:
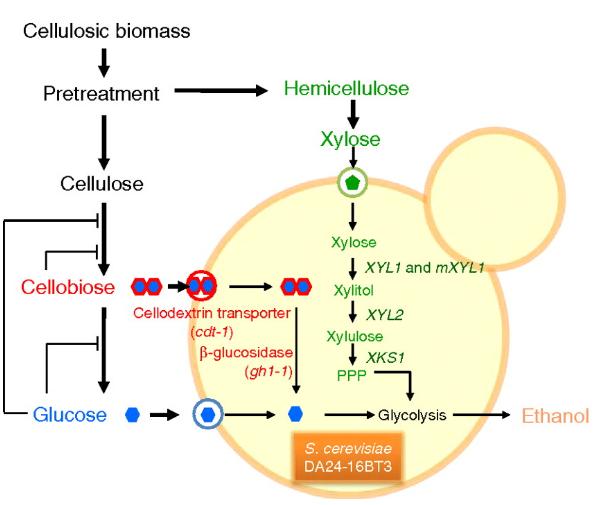
The efficient utilization of cellulose from plant material for biofuel production has been a popular subject in modern research. Cellulose contains many different hexoses (such as glucose) and pentoses (such as xylose), that can usually be fermented independently by various organisms, however the co-fermentation of the two has not been an efficient process thus far. In 2010, Suk-Jin Ha et al., worked to overcome this inefficiency by engineering a Saccharomyces cerevisiae to ferment both cellobiose (a glucose dimer) and xylose concurrently. Normally, S. cerevisiae that is used in ethanol biofuel production can efficiently ferment glucose as a carbon source, but not xylose since it lacks the necessary metabolic pathways The genes XYL1 and XYL2 encode the proteins xylose deductase (XR) and xylitol dehydrogenase (XDH) respectively, and the introduction of these genes into S. cerevisiae from Pichia stipitis can create a xylose hydrolysis pathway in the cell. However, now that the organism now had the ability to ferment both glucose and xylose, S. cerevisiae will still ferment glucose preferentially. In addition, once the glucose is depleted and a chance for xylose fermentation could occur, the ethanol levels are usually already so high that further fermentation is suppressed. To bypass this, Suk-Jin Ha et al also engineered a pathway into S. cerevisiae that would hydrolyze cellubiose instead of the glucose monomer. This was achieved by inserting the genes for a high affinity cellodextrin transporter, which is used for bringing cellubiose into the cell, and β-glucosidase, which hydrolyzes the cellubiose into glucose. Figure 5 displays the cofermentation of xylose and cellubiose in the cell. This engineered S. cerevisiae was shown to yield much higher levels of ethanol that S. cerevisiae that can only ferment glucose. This overall process makes the fermentation of plant cellulose into ethanol a much more efficient and useful method of biofuel production than it had ever been considered before 31.
Cool factor
Saccharomyces cerevisiae and Reactive Oxygen Species:
Typically when cells reduce oxygen during respiration, it occurs during the last step of the electron transport chain, producing hydrogen peroxide as a by-product 29. However, oxygen can also be reduced one step at a time using a single electron (an electron is transferred from a metal to an organic compound) 30. This pathway produces partially reduced end products such as the superoxide anion and hydrogen peroxide. These partially reduced end products are also known as reactive oxygen species or ROS 25.
ROS can react with most components of the cell, having damaging effects. In a normal, well functioning cell, the manufacturing and breaking down of ROS is controlled and regulated. However, sometimes the concentration of ROS can be higher than normal in a particular cell. When a cell is subjected to high ROS levels, it is called oxidative stress 24.
Radical oxidation of amino acids produce additional carbonyl groups in proteins. Carbonyl levels are used to measure how much ROS has damaged a protein 25. Amino acids containing sulfur, like tryptophan and tyrosine, are more prone to oxidation, and therefore the production of carbonyl groups 24.
In Saccharomyces cerevisiae, eliminating the Grx5 gene resulted in increased protein carbonyl levels and caused the proteins to be more susceptible to oxidation. Additionally, the molecular chaperone protein HSP60 activates protein folding in the mitochondrial matrix of cells 27. Cells of S. cerevisiae with higher HSP60 levels had fewer carbonyl groups in proteins than cells with lower levels of HSP60. HSP60 also protected the enzymes containing iron and sulfur from being deactivated 28. These enzymes prevent the spread of iron ions, which can be harmful to the cells. It has also been shown that higher and lower amounts of the superoxide dismutase (SOD) genes help to protect S. cerevisiae against the breaking down of proteins 26.
Therefore, S. cerevisiae has certain genes such as SOD, Grx5, and HSP60 that prevent the excess carbonylation of its proteins. So, it can be concluded that these genes help protect S. cerevisiae from ROS and by extension oxidative stress.
References
[2] White, Chris, and Jamil Zainasheff. Yeast: the Practical Guide to Beer Fermentation. Boulder, CO: Brewers Publications, 2010. Print.
[4] Madigan, Michael T. et al, 2009, Brock Biology of Microorganisms, San Francisco, CA, Pearson Education, Inc. Print.
[6] Saccharomyces Genome Database. 18 Feb. 2011. Web. 21 Feb. 2011. <http://www.yeastgenome.org/>.
[25] Jamieson, DJ. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast. 1998.
[29] Gregory, Michael J. Cellular Respiration. Clinton Community College.
Edited by the NC State University MB 103 class of 2007.
