Rhodospirillum rubrum: Difference between revisions
Cozzolino.m (talk | contribs) |
Cozzolino.m (talk | contribs) |
||
| (95 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
{{Biorealm Genus}} | {{Biorealm Genus}} | ||
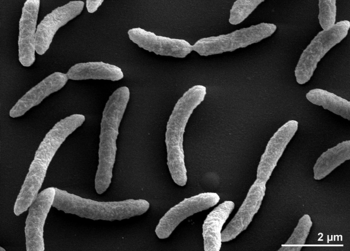
[[Image: | [[Image:rrubrums1.png|thumb|alt=R. rubrum|Rhodospirillum ruburm|350px|right|'''Image 1''' - ''Rhodospirillum rubrum'' strain S1T, SEM generated from a culture of DSM 467 (Source: Munk et al, 2011).]] | ||
==Classification== | ==Classification== | ||
| Line 17: | Line 17: | ||
''Rhodospirillum rubrum'' | ''Rhodospirillum rubrum'' | ||
==Description and significance== | ==Description and significance== | ||
''Rhodospirillum rubrum'' is a Gram-negative, mesophilicproteobacteria. Its optimal growth temperature is 25-30 degrees Celsius. It has multi-layered outer envelopes, which contain mostly unsaturated, but some saturated fats in its cell wall. ''R. rubrum'' is a spirilla, meaning it has a spiral-shape. It is polarly flagellated, and therefore motile. Its length is 3-10 um, with a width of | ''Rhodospirillum rubrum'' is a Gram-negative, mesophilicproteobacteria. Its optimal growth temperature is 25-30 degrees Celsius. It has multi-layered outer envelopes, which contain mostly unsaturated, but some saturated fats in its cell wall. ''R. rubrum'' is a spirilla, meaning it has a spiral-shape. It is polarly flagellated, and therefore motile. Its length is 3-10 um, with a width of 0.8-1.0 um. | ||
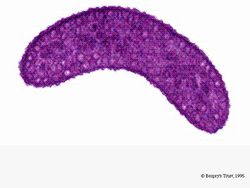
[[Image:rrubrum.jpg|thumb|alt=R. rubrum|Rhodospirillum ruburm| | [[Image:rrubrum.jpg|thumb|alt=R. rubrum|Rhodospirillum ruburm|250px|right|'''Image 2''' - ''Rhodospirillum ruburm'', TEM. Length: 4 microns (Copyright [http://4hgarden.msu.edu/kidstour/zoo/zwp0257.html Bergey's Manual Trust])]] | ||
''R. rubrum'' is a facultative anaerobe. Depending on the presence of oxygen, it can undergo alcoholic fermentation or aerobic respiration. It also is capable of photosynthesis and contains carotenoid and bateriochlorophyll in its chromatophore particles. These molecules help to absorb light and convert it to energy and also give it its distinct purple-red color under anaerobic conditions. ''R. rubrum'' is colorless under aerobic conditions. | ''R. rubrum'' is a facultative anaerobe. Depending on the presence of oxygen, it can undergo alcoholic fermentation or aerobic respiration. It also is capable of photosynthesis and contains carotenoid and bateriochlorophyll in its chromatophore particles. These molecules help to absorb light and convert it to energy and also give it its distinct purple-red color under anaerobic conditions. ''R. rubrum'' is colorless under aerobic conditions. | ||
Although photosynthesis is active under aerobic conditions, it is generally suppressed in the presence of | Although photosynthesis is active under aerobic conditions, it is generally suppressed in the presence of O<sub>2</sub>. Sulfur is a major byproduct of photosynthesis, not O<sub>2</sub>. ''R. rubrum'' can grow heterotrophically or autotrophically when photosynthetic. Unlike many plants, ''R. rubrum'' contains no chlorophyll a (absorption spectra 430-662 nm). However, it does contain chlorophyll b (absorption spectra 660-680 nm) and bacteriochlorophylls (800-925 nm). This allows it to utilize more energy from the electromagnetic spectra. ''R. rubrum'' also oxidizes carbon monoxide (CO) with hydrogen gas as the pathway’s end product, and can use sulfide at low concentrations as an electron donor in carbon dioxide reduction. Additionally, it is a nitrogen fixing bacteria; it uses nitrogenase to convert atmospheric nitrogen gas to ammonia (Munk et al, 2011). | ||
| Line 35: | Line 33: | ||
==Genome structure== | ==Genome structure== | ||
The genome's sequencing is finished. It consists of a circular chromosome with 4,352,825 base pairs, 65% of which are guanine-cystosine pairs. There is also a plasmid with 53,732 base pairs with 60% guanine-cytosine. In total, there are 3,850 protein coding genes and 83 RNA genes. Of these genes, 6.9% are transcription-related; 4.6% translation, ribosome structure, and biosynthesis; 4.0% replication, recombination and repair; 7.9% signal transduction mechanisms; 5.9% cell wall and membrane biogenesis; 6.6% energy production and conversion; 5.0% carbohydrate transport and metabolism; 9.9% amino acid transport and metabolism; 4.7% coenzyme transport and metabolism; 3.7% lipid transport and metabolism; and 6.5% inorganic ion transport and metabolism. | The genome's sequencing is finished. It consists of a circular chromosome with 4,352,825 base pairs, 65% of which are guanine-cystosine pairs. There is also a plasmid with 53,732 base pairs with 60% guanine-cytosine. In total, there are 3,850 protein coding genes and 83 RNA genes. Of these genes, 6.9% are transcription-related; 4.6% translation, ribosome structure, and biosynthesis; 4.0% replication, recombination and repair; 7.9% signal transduction mechanisms; 5.9% cell wall and membrane biogenesis; 6.6% energy production and conversion; 5.0% carbohydrate transport and metabolism; 9.9% amino acid transport and metabolism; 4.7% coenzyme transport and metabolism; 3.7% lipid transport and metabolism; and 6.5% inorganic ion transport and metabolism | ||
(Munk et al, 2011). | |||
==Ecology== | ==Ecology== | ||
[[Image:rrub2.jpg| | [[Image:rrub2.jpg|250px|thumb|right|'''Image 3''' - ''Rhodospirillum rubrum'', SEM. (Copyright [http://genome.jgi-psf.org/rhoru/rhoru.home.html Regents of the University of California])]] | ||
Due to the fact that ''Rhodospirillum rubrum'' can grow both aerobically and anaerobically, it is capable of inhabiting a wide variety of conditions. ''R. rubrum'' is found in many natural aquatic environments such as ponds, lakes, streams, and standing water (Reslewic et. al, 2005). ''R. rubrum'' is also often found in mud and sewage (Brock et al, 2000). Studies have shown that ''R. rubrum'' can make large changes in its chemical composition to adapt to different environments (Cohen-Bazire and Kunisawa, 1963). ''R. rubrum'' prefers to grow in habitats with a pH of 6.8-7.2 (Bergey and Holt, 1994). Studies have also shown that ''R. rubrum'' has an optimal growth temperature of 22-35 degrees Celcius (Weaver, 1971). | |||
==Cell structure and metabolism== | |||
''R. rubrum'' is a versatile organism that can obtain energy through a variety of mechanisms. Respiration and photosynthetic mechanisms exist together and their activity depends upon the presence of light and energy. ''R. rubrum'' can grow in dark chemo-tropical environments with the presence of oxygen or can grow in a photo tropical environment without oxygen. | |||
: | |||
Photosynthesis in ''R. rubrum'' begins with the donation of a hydrogen from an organic substrate to an oxidizing substrance. Bacteriochlorophyll and cartenoids, the photoreactive pigments found in the cell membrane, are bound to chromatophores (Schachman, Pardee and Stanier, 1952). Chromatophores are flattened disks that contain choline phospholipids, cardio lipin and galactosyl diglycerides (Benson, Wintermans, and Wiser, 1959; Benson 1961). Additionally, chromatophores contain a complete electron transport chain that includes various cytochromes, flavin and pyridine nucleotides (Newton and Newton, 1957; Hulcher and Conti, 1960; Kamen, 1961). The cell membrane also contains machinery for ATP synthesis, including an ATP synthetase. | |||
: | |||
In the presence of oxygen, ''R. rubrum'' is able to aerobically respire using a traditional electron transport chain with NAD+/NADH as the primary electron carrier (Keister and Minton, 1969). Among the electron transport proteins is cytochrome C428 (Chance and Smith, 1955). Aerobic respiration, however, is inhibited by photosynthesis (Oelze and Weaver, 1971). | |||
: | |||
''R. rubrum'' is also capable of anaerobic respiration. Its non-oxygenic terminal electron acceptors can include dimethyl sulfoxide and trimethylamine oxide. The presence of these electron acceptors makes it possible for substrates such as succinate, malate and acetate to support growth of ''R. rubrum''. These acceptors, however, are only approximately 33-41% as efficient as oxygen in terms of energy conservation (Schultz and Weaver, 1982). | |||
: | |||
Additionally, ''R. rubrum'' has the unique ability to oxidize carbon monoxide using carbon monoxide dehydrogenase. This oxidation pathway ends with the reduction and hydrogen and the production of hydrogen gas. | |||
: | |||
Without a terminal electron acceptor, ''R. rubrum'' completes mixed acid fermentation. The major products of pyruvate fermentation are acetate, formate, carbon dioxide and hydrogen. In the presence of bicarbonate ion, fructose is able to be fermented. The end products of this pathway are the same as that for pyruvate but with the addition of succinate and propionate (Schultz and Weaver 1982). | |||
: | |||
Besides having the ability to fix carbon dioxide (Schon and Biedermann, 1972), an identifying anabolic property of ''R. rubrum'' is its ability to fix nitrogen. Under dark conditions with the presence of fructose, ''R. rubrum'' uses nitrogenase to fix nitrogen gas to ammonium. It contains both Fe-Mo and Fe-only nitrogenases. | |||
==Pathology== | |||
[[Image:scope.jpg|thumb|alt=R. rubrum|Rhodospirillum ruburm|200px|right|'''Image 4''' - ''Rhodospirillum rubrum'', SEM. (Copyright [http://www.connecticutvalleybiological.com/rhodospirillum-rubrum-on-brain-heart-infusion-or-tryptic-soy-agar-tube-p-11180.html Connecticut Valley Biological Supply Co.])]] | |||
== | ''R. rubrum'' has not been found to infect humans or animals. | ||
==Current Research== | |||
===Radiation resistance=== | |||
''Rhodospirillum rubrum'' has recently provided insight into radiation resistance. Felice Mastroleo et al, in ''Insight into the radiotolerance of the life support bacterium Rhodospirillum rubrum S1H by means of phenotypic and transcriptomic methods'' (2010), have characterized the radiation tolerance of ''R. rubrum''. | |||
By subjecting the bacteria to sub-lethal doses of Cobalt-60 gamma rays, a type of ionizing radiation, ''R. rubrum'' marker genes specific to ionizing radiation were characterized in the organism. These genes increased in expression as "recovery time" elapsed after a given gamma radiation treatment. The analysis of gene expression by ''R. rubrum'' post-gamma radiation creates the potential for further microbiological research that may lead to advances in applied radiation resistance in the future. | |||
===Biodegradable plastics=== | |||
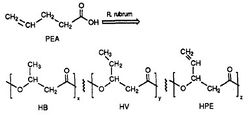
[[Image: | [[Image:plastic.jpg|thumb|alt=R. rubrum|Rhodospirillum ruburm|250px|right|'''Figure 1''' - ''Rhodospirillum rubrum'': synthesis of the biodegradable polyester, PHA (Source: Ulmer et al, 1993).]] | ||
''R. rubrum'' is found to produce a class of biodegradable plastics, poly beta-hydroxyalkanoates (PHAs). The applications of this research can solve many problems caused by synthetic, non-degradable plastics that have adversely impacted the environment. A research experiment performed by Herbert. W. Ulmer et al chronicled the production of PHAs containing 3HV units by ''Rhodospriillum rubrum'' (1993). Ulmer used 4-pentenoic acid (PEA) as the growth medium for ''R. rubrum''. When the bacteria was placed under "PHA-producing conditions," PHAs were synthesized containing 3 repeating subunits (note: R = substituent group): HB (R = methyl), HV (R = ethyl), HPE (R = vinyl) (See Figure 1). | |||
The synthesis of functional, biodegradable plastics by bacteria has the potential to reduce global pollution and fuel expenditure from plastic factories. ''R. rubrum'' can be used as a microbiological tool to produce eco-friendly biopolymers. | |||
===Renewable energy=== | |||
The rising concern over environmental pollution from burning fossil fuels has led to an increase in research of ''Rhodospirillum rubrum''. Using ''R. rubrum'' as a source for hydrogen to be used in renewable hydrogen fuels addresses several problems created by fossil fuel consumption. Hydrogen oxidizes to water as a combustion product, unlike fossil fuels that create pollutants such as CO, CO<sub>2</sub>, SO<sub>x</sub>, and NO<sub>x</sub>. ''R. rubrum'' can produce hydrogen by utilizing the following "water-gas shift reaction" (Najafpour et al, 2004): | |||
:::CO + H<sub>2</sub>O → H<sub>2</sub> + CO<sub>2</sub> | |||
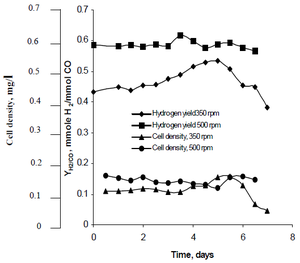
[[Image:najafpour.png|thumb|alt=R. rubrum|Rhodospirillum ruburm|300px|right|'''Figure 2''' - Effect of agitation rate on cell density and hydrogen production in continuous fermentation (Source: Najafpour et al, 2004).]] | |||
Najafpour's research method observed how agitation, created by sets of turbine impellers set at rates of 350 rpm as well as 500 rpm, affected the hydrogen production rates of ''R. rubrum''. His research found that hydrogen fuel has the potential to curb global warming, and provide an eco-friendly technological solution to fuel production and consumption. His study revealed "higher agitation rate yielded higher hydrogen as well as cell density" of ''R. rubrum'' (See Figure 2). | |||
In another microbiological study, performed by Matthew Ross Melnicki, it was found that ''R. rubrum'' provides the potential for renewable energy, such as hydrogen fuel, via hydrogen synthesis. Its biological pathways can produce hydrogen levels at high rates when co-cultivated with ''Chlamydomonas reinhardtii'', an algal species which ''R. rubrum'' can grow harmoniously with. Melnicki proposed an unconventional co-cultivation approach to synthesizing hydrogen that produced enhanced levels of hydrogen synthesis that may not be achieved if the co-cultivation is uncoupled. ''C. reinhardtii'' was favored at higher light intensities, while ''R. ruburm'' was more productive under lower light intensities, highlighting the advantages of their co-cultivation. | |||
By using a fed-batch technique for cultivating ''R. rubrum'', it was found that inoculated cultures produced hydrogen during its exponential growth phase, and the synthesis of hydrogen continued to flourish continuously for 70 hours. When growth ceased, Melnicki added succinate, which acts as an electron donor in the metabolic pathway, and hydrogen evolution resumed even though the bacteria now longer grew. However, it is important to note that with continual addition of succinate, reduced evolution rates of hydrogen were found. Once additional growth media was added, rates of hydrogen production increased once again. | |||
Melnicki also stated, "while growth is not required for hydrogen production, this work establishes the necessity for cell growth in order to maintain maximal rates, suggesting the industrial suitability of a semi-continuous culture strategy" (Melnicki, 2009). | |||
== | ==Cool Factor== | ||
The genome sequence of ''Rhodospirillum rubrum'' is only the second of the ''Rhodospirillum'' genus that has been published, after ''R. centenum'' (Munk et al, 2011). | |||
[[Image:rrubrumpurple.jpg|thumb|alt=R. rubrum|Rhodospirillum ruburm|250px|right|'''Image 5''' - ''Rhodospirillum rubrum'' under light microscopy (Copyright [http://www.corbisimages.com/stock-photo/rights-managed/42-20490851/rhodospirillum-rubrum-bacteria Corbis])]] | |||
An interesting aspect of ''Rhodospirillum rubrum'' is its ability to change color under different conditions. It is an anoxygenic phototroph, but does not produce extracellular oxygen. Instead, it produces elemental sulfur during light harvesting. This is due to the fact that ''R. rubrum'' lacks light harvesting complex 2 (Munk et al, 2011). The bacterium uses bacteriochlorophyll, enabling it to absorb wavelengths of light that plants do not utilize. Bacteriochlorophyll absorbs wavelengths 800-925 nm. | |||
Under aerobic conditions, photosynthetic activity of ''R. rubrum'' is genetically surpressed, and the bacteria becomes colorless. Under anaerobic conditions, however, photosynthesis is activated and the bacteria takes on a distinct purple-red color (see Image 5). This is because due to activity in the chromatophores, found in the membrane (Munk et al, 2011). Chromatophores contain the bacterium's bacteriochlorophyll pigments, and undergo absorption changes during photosynthesis activation. | |||
==References== | |||
Benson, A. “Chloroplast lipid metabolism.” In Symposium no. VI. 5th international Congress on Biochemistry. 1961. Oxford: Pergamon Press. (In the Press.) | |||
Benson, A., Wintermans, J. and Wiser, R. “Chloroplast lipids as carbohydrate reservoirs.” Plant Physiology. 1959. Volume 34. p. 815. | |||
Bergey, D. and Holt, J. Bergey’s manual of determinative bacteriology. Ninth edition. Baltimore. Lippincott Williams and Wilkins. 1994. p. 359. | |||
Brock, T.D., M.T. Madigan, J.M. Martinko, and J Parker. Biology of Microorganisms. 9th Edition. Upper Saddle River: Prentice Hall, 2000. p. 459. | |||
Chance, B. and Smith, L. “Respiratory pigments of Rhodospirillum rubrum.” Nature. Volume 174. p. 803-806. | |||
Cohen-Bazire, G. and Kunisawa, R. "The Fine Structure of Rhodospirillum Rubrum ." Journal of Cell Biology. 1963. Volume 16. p. 401-419. | |||
Hulcher, M. and Conti, S. “Cytochronies in chlorophyll-containing particles of Chromatium and Chlorobium thiosuphatophilum.” Biochemistry and Biophysics Research Communications. 1960. Volume 3. p. 497. | |||
Kamen, M. “The Haem Proteins of Photosynthetic Tissues.” In Symposium no. VI. 5th international Congress on Biochemistry. 1961. Oxford: pergamon Press. (In the Press.) | |||
Keister, D. and Minton, N. “Energy-linked reactions in photosynthetic bacteria. 3. Further studies on energy-linked nicotinamide-adenine dinucleotide reduction by Rhodospirillum rubrum chromatophores.” Biochemistry. 1969. Volume 8. p. 167-173. | |||
[http://adsabs.harvard.edu/abs/2010cosp...38.3127M Mastroleo, F., Monsieurs, P., and Leys, N. "''Insight into the radiotolerance of the life support bacterium Rhodospirillum rubrum S1H by means of phenotypic and transcriptomic methods". ''COSPAR Scientific Assembly''. 2010. p. 4.] | |||
[http://epmb.berkeley.edu/vfs/PPs/Melnicki-MatR/web/MMCV.pdf Melnicki, M. R. "''Modes of hydrogen production in the photosynthetic bacterium, Rhodosphirillum rubrum". ''University of California, Berkeley''. 2009.] | |||
[http://standardsingenomics.org/index.php/sigen/article/viewArticle/sigs.1804360 Munk, C., Copeland, A., Lucas, C., Lapidus, A., et al. "''Complete genome sequence of Rhodospirillum rubrum type strain (S1T)". ''Standards in Genomic Sciences''. 2011. Volume 4. pp. 293-302.] | |||
[http://www.ajol.info/index.php/ajb/article/viewFile/15006/58911 Najafpour, G., Ismail, K., Younesi, H., Mohamed, A. R., and Kamaruddin, A. H. "''Hydrogen as clean fuel via continuous fermentation by anaerobic photosynthetic bacteria, Rhodospirillum rubrum". ''African Journal of Biotechnology''. 2004. Volume 3. pp. 503-507.] | |||
Newton, J. and Newton, G. “Composition of the photoactive subcellular particles from Chromatium.” The Archives of Biochemistry and Biophysics. 1957. Volume 71. p. 250. | |||
Oelze, J. and Weaver, P. “The adjustment of photosynthetically grown cells of Rhodospirillum rubrum to aerobic light conditions.” The Archives of Microbiology. 1982. Volume 79. p. 108-121. | |||
Reslewic, S., Shigou, Z., Place, M., Zhang, Y., Briska A., Goldstein, S., Churas, C., Runnheim, R., Forrest, D., Lim, A., Lapidus, A., Han, C., Roberts, G., and Schwartz, D. "Whole-Genome Shotgun Optical Mapping of Rhodospirillum rubrum ." Applied and Environmental Microbiology. 2005. Volume 71. p. 5511-5522. | |||
Schachman, H., Pardee, A., and Stanier, R. “Studies on the macromolecular organization of microbial cells.” The Archives of Biochemistry and Biophysics. 1952. Volume 38. p. 245-260. | |||
Schön, G. and Biedermann, M. “Synthesis of volatile acids by fermentation of pyruvate and fructose in anaerobic dark cultures of Rhodospirillum rubrum.” Archiv fur Mikrobiologie.1972. Volume 85. p. 77-90. | |||
Schultz, J. and Weaver, P. “Fermentation and anaerobic respiration by Rhodospirillum rubrum and Rhodopseudomonas capsulate.” The Journal of Bacteriology. 1982. Volume 149. p. 181-190. | |||
[http://pubs.acs.org/doi/abs/10.1021/ma00085a002 Ulmer, H., Gross, R., Posada, M., Weisbach, P., Fuller, R. C., and Lenz, R. "''Bacterial production of poly(.beta.-hydroxyalkanoates) containing unsaturated repeating units by Rhodospirillum rubrum". ''Macromolecules''. 1994. Volume 27. p. 1675-1679.] | |||
Weaver, P. “Temperature-Sensitive Mutations of the Photosynthetic Apparatus of Rhodospirillum rubrum." Proceedings of the National Academy of Sciences of the United States of America. 1971. Volume 68. p. 136-138. | |||
'''Edited by students of Dr. Iris Keren: Allison Almeida, Matthew Cozzolino, Trung Nguyen, and Jessica Wong.''' | |||
NEUF2011 | |||
Latest revision as of 18:41, 3 November 2011
A Microbial Biorealm page on the genus Rhodospirillum rubrum
Classification
Higher order taxa
Kingdom: Bacteria
Phylum: Proteobacteria
Class: Alphaproteobacteria
Order: Rhodospirillales
Family: Rhodospirillaceae
Genus: Rhodospirillum
Species
Rhodospirillum rubrum
Description and significance
Rhodospirillum rubrum is a Gram-negative, mesophilicproteobacteria. Its optimal growth temperature is 25-30 degrees Celsius. It has multi-layered outer envelopes, which contain mostly unsaturated, but some saturated fats in its cell wall. R. rubrum is a spirilla, meaning it has a spiral-shape. It is polarly flagellated, and therefore motile. Its length is 3-10 um, with a width of 0.8-1.0 um.
R. rubrum is a facultative anaerobe. Depending on the presence of oxygen, it can undergo alcoholic fermentation or aerobic respiration. It also is capable of photosynthesis and contains carotenoid and bateriochlorophyll in its chromatophore particles. These molecules help to absorb light and convert it to energy and also give it its distinct purple-red color under anaerobic conditions. R. rubrum is colorless under aerobic conditions.
Although photosynthesis is active under aerobic conditions, it is generally suppressed in the presence of O2. Sulfur is a major byproduct of photosynthesis, not O2. R. rubrum can grow heterotrophically or autotrophically when photosynthetic. Unlike many plants, R. rubrum contains no chlorophyll a (absorption spectra 430-662 nm). However, it does contain chlorophyll b (absorption spectra 660-680 nm) and bacteriochlorophylls (800-925 nm). This allows it to utilize more energy from the electromagnetic spectra. R. rubrum also oxidizes carbon monoxide (CO) with hydrogen gas as the pathway’s end product, and can use sulfide at low concentrations as an electron donor in carbon dioxide reduction. Additionally, it is a nitrogen fixing bacteria; it uses nitrogenase to convert atmospheric nitrogen gas to ammonia (Munk et al, 2011).
There are several applications of R. rubrum in the field of biotechnology. It is a model system of light to chemical energy conversion and for its nitrogen fixing pathways. It is also the subject of radiation resistance studies. It can be used in several ways for consumption, as well. The proteobacteria may be a source of animal food and agricultural fertilizer. Another important role in research includes the production of vitamins. It is also being researched for its production of biological plastic from precursors of poly-hydroxy-butric-acid. R. rubrum may also be a contributor in biological hydrogen fuels, mainly through its evolution of the enzyme nitrogenase.
Genome structure
The genome's sequencing is finished. It consists of a circular chromosome with 4,352,825 base pairs, 65% of which are guanine-cystosine pairs. There is also a plasmid with 53,732 base pairs with 60% guanine-cytosine. In total, there are 3,850 protein coding genes and 83 RNA genes. Of these genes, 6.9% are transcription-related; 4.6% translation, ribosome structure, and biosynthesis; 4.0% replication, recombination and repair; 7.9% signal transduction mechanisms; 5.9% cell wall and membrane biogenesis; 6.6% energy production and conversion; 5.0% carbohydrate transport and metabolism; 9.9% amino acid transport and metabolism; 4.7% coenzyme transport and metabolism; 3.7% lipid transport and metabolism; and 6.5% inorganic ion transport and metabolism (Munk et al, 2011).
Ecology
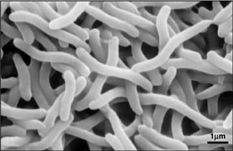
Due to the fact that Rhodospirillum rubrum can grow both aerobically and anaerobically, it is capable of inhabiting a wide variety of conditions. R. rubrum is found in many natural aquatic environments such as ponds, lakes, streams, and standing water (Reslewic et. al, 2005). R. rubrum is also often found in mud and sewage (Brock et al, 2000). Studies have shown that R. rubrum can make large changes in its chemical composition to adapt to different environments (Cohen-Bazire and Kunisawa, 1963). R. rubrum prefers to grow in habitats with a pH of 6.8-7.2 (Bergey and Holt, 1994). Studies have also shown that R. rubrum has an optimal growth temperature of 22-35 degrees Celcius (Weaver, 1971).
Cell structure and metabolism
R. rubrum is a versatile organism that can obtain energy through a variety of mechanisms. Respiration and photosynthetic mechanisms exist together and their activity depends upon the presence of light and energy. R. rubrum can grow in dark chemo-tropical environments with the presence of oxygen or can grow in a photo tropical environment without oxygen.
Photosynthesis in R. rubrum begins with the donation of a hydrogen from an organic substrate to an oxidizing substrance. Bacteriochlorophyll and cartenoids, the photoreactive pigments found in the cell membrane, are bound to chromatophores (Schachman, Pardee and Stanier, 1952). Chromatophores are flattened disks that contain choline phospholipids, cardio lipin and galactosyl diglycerides (Benson, Wintermans, and Wiser, 1959; Benson 1961). Additionally, chromatophores contain a complete electron transport chain that includes various cytochromes, flavin and pyridine nucleotides (Newton and Newton, 1957; Hulcher and Conti, 1960; Kamen, 1961). The cell membrane also contains machinery for ATP synthesis, including an ATP synthetase.
In the presence of oxygen, R. rubrum is able to aerobically respire using a traditional electron transport chain with NAD+/NADH as the primary electron carrier (Keister and Minton, 1969). Among the electron transport proteins is cytochrome C428 (Chance and Smith, 1955). Aerobic respiration, however, is inhibited by photosynthesis (Oelze and Weaver, 1971).
R. rubrum is also capable of anaerobic respiration. Its non-oxygenic terminal electron acceptors can include dimethyl sulfoxide and trimethylamine oxide. The presence of these electron acceptors makes it possible for substrates such as succinate, malate and acetate to support growth of R. rubrum. These acceptors, however, are only approximately 33-41% as efficient as oxygen in terms of energy conservation (Schultz and Weaver, 1982).
Additionally, R. rubrum has the unique ability to oxidize carbon monoxide using carbon monoxide dehydrogenase. This oxidation pathway ends with the reduction and hydrogen and the production of hydrogen gas.
Without a terminal electron acceptor, R. rubrum completes mixed acid fermentation. The major products of pyruvate fermentation are acetate, formate, carbon dioxide and hydrogen. In the presence of bicarbonate ion, fructose is able to be fermented. The end products of this pathway are the same as that for pyruvate but with the addition of succinate and propionate (Schultz and Weaver 1982).
Besides having the ability to fix carbon dioxide (Schon and Biedermann, 1972), an identifying anabolic property of R. rubrum is its ability to fix nitrogen. Under dark conditions with the presence of fructose, R. rubrum uses nitrogenase to fix nitrogen gas to ammonium. It contains both Fe-Mo and Fe-only nitrogenases.
Pathology
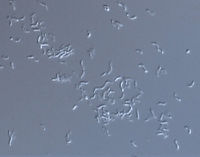
R. rubrum has not been found to infect humans or animals.
Current Research
Radiation resistance
Rhodospirillum rubrum has recently provided insight into radiation resistance. Felice Mastroleo et al, in Insight into the radiotolerance of the life support bacterium Rhodospirillum rubrum S1H by means of phenotypic and transcriptomic methods (2010), have characterized the radiation tolerance of R. rubrum.
By subjecting the bacteria to sub-lethal doses of Cobalt-60 gamma rays, a type of ionizing radiation, R. rubrum marker genes specific to ionizing radiation were characterized in the organism. These genes increased in expression as "recovery time" elapsed after a given gamma radiation treatment. The analysis of gene expression by R. rubrum post-gamma radiation creates the potential for further microbiological research that may lead to advances in applied radiation resistance in the future.
Biodegradable plastics
R. rubrum is found to produce a class of biodegradable plastics, poly beta-hydroxyalkanoates (PHAs). The applications of this research can solve many problems caused by synthetic, non-degradable plastics that have adversely impacted the environment. A research experiment performed by Herbert. W. Ulmer et al chronicled the production of PHAs containing 3HV units by Rhodospriillum rubrum (1993). Ulmer used 4-pentenoic acid (PEA) as the growth medium for R. rubrum. When the bacteria was placed under "PHA-producing conditions," PHAs were synthesized containing 3 repeating subunits (note: R = substituent group): HB (R = methyl), HV (R = ethyl), HPE (R = vinyl) (See Figure 1).
The synthesis of functional, biodegradable plastics by bacteria has the potential to reduce global pollution and fuel expenditure from plastic factories. R. rubrum can be used as a microbiological tool to produce eco-friendly biopolymers.
Renewable energy
The rising concern over environmental pollution from burning fossil fuels has led to an increase in research of Rhodospirillum rubrum. Using R. rubrum as a source for hydrogen to be used in renewable hydrogen fuels addresses several problems created by fossil fuel consumption. Hydrogen oxidizes to water as a combustion product, unlike fossil fuels that create pollutants such as CO, CO2, SOx, and NOx. R. rubrum can produce hydrogen by utilizing the following "water-gas shift reaction" (Najafpour et al, 2004):
- CO + H2O → H2 + CO2
Najafpour's research method observed how agitation, created by sets of turbine impellers set at rates of 350 rpm as well as 500 rpm, affected the hydrogen production rates of R. rubrum. His research found that hydrogen fuel has the potential to curb global warming, and provide an eco-friendly technological solution to fuel production and consumption. His study revealed "higher agitation rate yielded higher hydrogen as well as cell density" of R. rubrum (See Figure 2).
In another microbiological study, performed by Matthew Ross Melnicki, it was found that R. rubrum provides the potential for renewable energy, such as hydrogen fuel, via hydrogen synthesis. Its biological pathways can produce hydrogen levels at high rates when co-cultivated with Chlamydomonas reinhardtii, an algal species which R. rubrum can grow harmoniously with. Melnicki proposed an unconventional co-cultivation approach to synthesizing hydrogen that produced enhanced levels of hydrogen synthesis that may not be achieved if the co-cultivation is uncoupled. C. reinhardtii was favored at higher light intensities, while R. ruburm was more productive under lower light intensities, highlighting the advantages of their co-cultivation.
By using a fed-batch technique for cultivating R. rubrum, it was found that inoculated cultures produced hydrogen during its exponential growth phase, and the synthesis of hydrogen continued to flourish continuously for 70 hours. When growth ceased, Melnicki added succinate, which acts as an electron donor in the metabolic pathway, and hydrogen evolution resumed even though the bacteria now longer grew. However, it is important to note that with continual addition of succinate, reduced evolution rates of hydrogen were found. Once additional growth media was added, rates of hydrogen production increased once again.
Melnicki also stated, "while growth is not required for hydrogen production, this work establishes the necessity for cell growth in order to maintain maximal rates, suggesting the industrial suitability of a semi-continuous culture strategy" (Melnicki, 2009).
Cool Factor
The genome sequence of Rhodospirillum rubrum is only the second of the Rhodospirillum genus that has been published, after R. centenum (Munk et al, 2011).

An interesting aspect of Rhodospirillum rubrum is its ability to change color under different conditions. It is an anoxygenic phototroph, but does not produce extracellular oxygen. Instead, it produces elemental sulfur during light harvesting. This is due to the fact that R. rubrum lacks light harvesting complex 2 (Munk et al, 2011). The bacterium uses bacteriochlorophyll, enabling it to absorb wavelengths of light that plants do not utilize. Bacteriochlorophyll absorbs wavelengths 800-925 nm.
Under aerobic conditions, photosynthetic activity of R. rubrum is genetically surpressed, and the bacteria becomes colorless. Under anaerobic conditions, however, photosynthesis is activated and the bacteria takes on a distinct purple-red color (see Image 5). This is because due to activity in the chromatophores, found in the membrane (Munk et al, 2011). Chromatophores contain the bacterium's bacteriochlorophyll pigments, and undergo absorption changes during photosynthesis activation.
References
Benson, A. “Chloroplast lipid metabolism.” In Symposium no. VI. 5th international Congress on Biochemistry. 1961. Oxford: Pergamon Press. (In the Press.)
Benson, A., Wintermans, J. and Wiser, R. “Chloroplast lipids as carbohydrate reservoirs.” Plant Physiology. 1959. Volume 34. p. 815.
Bergey, D. and Holt, J. Bergey’s manual of determinative bacteriology. Ninth edition. Baltimore. Lippincott Williams and Wilkins. 1994. p. 359.
Brock, T.D., M.T. Madigan, J.M. Martinko, and J Parker. Biology of Microorganisms. 9th Edition. Upper Saddle River: Prentice Hall, 2000. p. 459.
Chance, B. and Smith, L. “Respiratory pigments of Rhodospirillum rubrum.” Nature. Volume 174. p. 803-806.
Cohen-Bazire, G. and Kunisawa, R. "The Fine Structure of Rhodospirillum Rubrum ." Journal of Cell Biology. 1963. Volume 16. p. 401-419.
Hulcher, M. and Conti, S. “Cytochronies in chlorophyll-containing particles of Chromatium and Chlorobium thiosuphatophilum.” Biochemistry and Biophysics Research Communications. 1960. Volume 3. p. 497.
Kamen, M. “The Haem Proteins of Photosynthetic Tissues.” In Symposium no. VI. 5th international Congress on Biochemistry. 1961. Oxford: pergamon Press. (In the Press.)
Keister, D. and Minton, N. “Energy-linked reactions in photosynthetic bacteria. 3. Further studies on energy-linked nicotinamide-adenine dinucleotide reduction by Rhodospirillum rubrum chromatophores.” Biochemistry. 1969. Volume 8. p. 167-173.
Newton, J. and Newton, G. “Composition of the photoactive subcellular particles from Chromatium.” The Archives of Biochemistry and Biophysics. 1957. Volume 71. p. 250.
Oelze, J. and Weaver, P. “The adjustment of photosynthetically grown cells of Rhodospirillum rubrum to aerobic light conditions.” The Archives of Microbiology. 1982. Volume 79. p. 108-121.
Reslewic, S., Shigou, Z., Place, M., Zhang, Y., Briska A., Goldstein, S., Churas, C., Runnheim, R., Forrest, D., Lim, A., Lapidus, A., Han, C., Roberts, G., and Schwartz, D. "Whole-Genome Shotgun Optical Mapping of Rhodospirillum rubrum ." Applied and Environmental Microbiology. 2005. Volume 71. p. 5511-5522.
Schachman, H., Pardee, A., and Stanier, R. “Studies on the macromolecular organization of microbial cells.” The Archives of Biochemistry and Biophysics. 1952. Volume 38. p. 245-260.
Schön, G. and Biedermann, M. “Synthesis of volatile acids by fermentation of pyruvate and fructose in anaerobic dark cultures of Rhodospirillum rubrum.” Archiv fur Mikrobiologie.1972. Volume 85. p. 77-90.
Schultz, J. and Weaver, P. “Fermentation and anaerobic respiration by Rhodospirillum rubrum and Rhodopseudomonas capsulate.” The Journal of Bacteriology. 1982. Volume 149. p. 181-190.
Weaver, P. “Temperature-Sensitive Mutations of the Photosynthetic Apparatus of Rhodospirillum rubrum." Proceedings of the National Academy of Sciences of the United States of America. 1971. Volume 68. p. 136-138.
Edited by students of Dr. Iris Keren: Allison Almeida, Matthew Cozzolino, Trung Nguyen, and Jessica Wong.
NEUF2011