Haemobartonella felis: Difference between revisions
Maggiebenson (talk | contribs) No edit summary |
Maggiebenson (talk | contribs) |
||
| (116 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
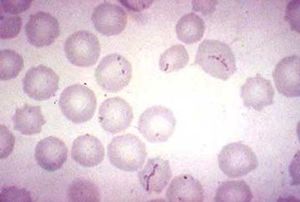
[[File:Img0027.jpg |thumb| Figure 1. Cell Morphology for ''Haemobartonella felis'' [http://instruction.cvhs.okstate.edu/jcfox/htdocs/disk1/images/img0027.jpg]]] | |||
''[[Haemobartonella felis]]'' | |||
==Characteristics of the symbiont/pathogen== | ==Characteristics of the symbiont/pathogen== | ||
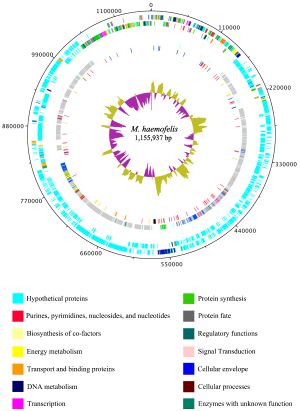
''Haemobartonella felis'' has been recently renamed to ''Mycoplasma haemofelis'' due to a closer relation to the genus ''Mycoplasma'' rather than the genus ''Bartonella'' (10). The genome of ''M. haemofelis'' has been sequenced, and in fact is the first hemotrophic mycoplasma species to be completely sequenced. In a study that compared two strains of the microbe, Ohio2 and Langford 1, they found that the complete genome of the circular chromosome was 1,155,937 base pairs. Even though the genome of ''M. haemofelis'' is small like other mycoplasmas, it still contains a large number of unique proteins (Figure 2). This large set of proteins is most likely due to its target for red blood cells, which may require a specific set of genes (5). | |||
The phylogenetic classification of ''M. haemofelis'' is (4): | |||
:Domain: Bacteria | |||
:Phylum: Tenericutes | |||
:Class: Mollicutes | |||
:Family: Mycoplasmataceae | |||
:Genus: Mycoplasma | |||
:Species: Haemofelis | |||
The cell morphology of ''M. haemofelis'' is a short coccus- or rod-shaped , gram negative bacteria. It also lacks a cell wall. Because the microbe is unable to be cultured,a microscopic identification from a blood smear is needed for diagnosis (3). | |||
[[File:1297-9716-42-102-1.jpg|thumb|Figure 2. Structure of the genome of ''Mycoplasma haemofelis''[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3196708/figure/F1/]]] | |||
==Characteristics of the host== | ==Characteristics of the host== | ||
The hosts for ''Mycoplasma haemofelis'' are felines. It is unclear of what breeds of felines are most commonly effected. However, it is found that felines of older non-pedigree breeds are more at risk for the infection that ''M. haemofelis'' even though younger felines can be affected (9). In other studies, they found that the infection is more common in felines from regions like Switzerland and other European countries (11). | |||
==Host-Symbiont Interaction == | ==Host-Symbiont Interaction == | ||
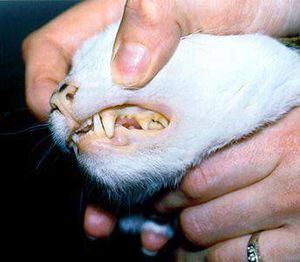
The interaction of the host, a cat, and the symbiont, ''Mycoplasma haemofelis'', is one of a parasitic relationship. If the host acquires a relationship with the symbiont, the host will become anemic, which affects the host's red blood cell count. It is unclear of how the symbiont and disease is transmitted. However, some studies suggest that it is transmitted by fleas, so flea control is suggested for felines. Because it is known that the symbiont causes problems in the host's blood, it can be determined that another source of transmission is by blood, such as from blood donor cats that are in areas where the infection occurs (7). One of the symptoms for this disease are pale gums (Figure 3). | |||
Since transmission is unclear, treatments are possible but not known to completely treat anemia for the infected feline. The most commonly used treatment is doxycycline that can be used for 14-21 days with one dosage a day. If the anemic conditions of the host are severe, a blood transfusion may be recommended (7). | |||
The interaction is obligate, because the microbe ''M. haemofelis'' needs the host in order to carry out specific cellular functions. The class of ''M. haemofelis'','' Mollicutes'', have been studied and determined to be dependent on hosts or other bacterium for guanine phosphoribosyl-transferase activity in order to carry out purine metabolism (9). | |||
[[File:Infosheet anaemia 1.jpg|thumb|Figure 3. A feline infected by ''Mycoplasma haemofelis'' that has caused pale gums.[http://www.fabcats.org/owners/anaemia/info.html]]] | |||
==Molecular Insights into the Symbiosis== | ==Molecular Insights into the Symbiosis== | ||
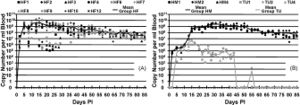
In some recent studies, the species ''Mycoplasma haemofelis'' has been compared to two other haemoplasma species, ''Candidatus'' M. haemominutum and ''Candidatus'' M. turicensis. In comparison of effects on the host of the symbiont, ''M. haemofelis'' causes haemolysis and ''C.'' M. haemominutum does not usually cause anemia. Coombs' tests and blood glucose concentration tests were used to determine the differences between these three species. Coomb's testing determines the presence for erythrocyte-bound antibodies and if the red blood cells become destroyed. The species ''M. haemofelis'' was the only species that tested positive for Coomb's test. In addition, the species ''C.'' M. turnicensis had lower glucose concentrations than the other two species as well as lower blood copy numbers (Figure 4). In conclusion of this study, they found that the species ''M. haemofelis'' was the only to develop macrocytic anemia (6). | |||
Many genes of ''M. haemofelis'' have been studied. The two genes IMP (inosine-5′-monophosphate) dehydrogenase and GMP (guanosine 3′,5′-monophosphate) synthase have been found to encode proteins and partake in purine biosynthesis. This is significant because these enzymatic activities of these genes have not been found in ''Mollicutes''. This raises the question if ''M. haemofelis'' needs a host in order to perform guanine biosynthesis,because it lacks the cellular components and functions for the biosynthesis. However, there is not enough research completed in order to determine this. Also, it is not clear what are the enzymes activities. In addition, the gene superoxide dismutase (SOD) is very unique to ''M. haemofelis'' because it is not found or sequenced in other mycoplasmas. However, there is no clear evidence of the genes' activity. Furthermore, it is found that this specie has many unique proteins and genes. Because of the unique genes, it is understood that this species of mycoplasma has adapted it self to colonize the red blood cell rather than mucosal surfaces like most mycoplasmas do and this is why it survives in its host (2). | |||
[[File:Tasker blood count.jpg|thumb|Figure 4. Haemoplasma copy numbers from Tasker (2009) study. (A) ''M. haemofelis'' copy number/ ml of blood for each cat versus time. (B) ''C.'' M. haemominutum and ''C.'' M. turnicensis. Means are plotted as continuous line.''C.'' M. turnicensis had lower copy numbers than other two groups.[http://www.sciencedirect.com/science?_ob=MiamiCaptionURL&_method=retrieve&_eid=1-s2.0-S0378113509003137&_image=1-s2.0-S0378113509003137-gr1.jpg&_ba=&_fmt=full&_orig=na&_issn=03781135&_pii=S0378113509003137&_acct=C000069779&_version=1&_urlVersion=0&_userid=6406178&md5=ea21e24d2afd0ed4ab92cdae29e1543a]]] | |||
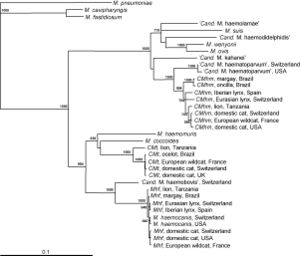
[[File:Willi phylogenetic map.jpg|thumb|Figure 5. Phylogenetic analysis of ''Mycoplasma haemofelis'' to ''Candidatus'' M. haemominutum and ''Candidatus'' M. turnicensis[http://www.sciencedirect.com/science?_ob=MiamiCaptionURL&_method=retrieve&_eid=1-s2.0-S0378113507003240&_image=1-s2.0-S0378113507003240-gr1.jpg&_ba=&_fmt=full&_orig=na&_issn=03781135&_pii=S0378113507003240&_acct=C000069779&_version=1&_urlVersion=0&_userid=6406178&md5=bc603d968e1825f510a47315b44bf208]]] | |||
==Ecological and Evolutionary Aspects == | ==Ecological and Evolutionary Aspects == | ||
In the past decade or so, ''Mycoplasma haemofelis'' has been difficult to diagnosis. However, with more recent research it is possible to diagnose for the infection by the polymerase chain reaction (PCR) rather than the older method of studying the red blood cells, which resulted in false positive and false negative due to other factors. These new PCR methods have contributed to the identification of two strains of ''M. haemofelis'', Ohio strain and California strain. The Ohio strain causes a more severe infection than California strain and it could result in haemolytic anemia that could become fatal. The fact that strains are from different geological regions and that most felines affected are found in Europe may suggest that environmental factors play a role in regulating the symbiosis. However, ''M. haemofelis'' is still in the early stages of research because of its difficulty in culturing, so as of now environmental factors that regulate the parthenogenesis are not determined (7). | |||
==Recent Discoveries== | ==Recent Discoveries== | ||
In one study, the ''Mycoplasma haemofelis'' strain Ohio2 was used for genome sequencing in order to understand why the symbiont needs the host for metabolic pathways and resources in order to survive. ''M. haemofelis'' relies on its host because it lacks necessities for some metabolisms, such as pyrimidines and amino acids. However, this study represents some of the differences that ''M. haemofelis'' uses in some other metabolisms. For example, mycoplasmas have been thought to be incapable of fatty acid biosynthesis from acetyl-CoA because of a lack of genetic material. Yet, this study finds that ''M. haemofelis'' could be able to use glycerol in order to synthesize phospholipids. In addition, this study identified two virulence genes for ''M. haemofelis''. One of these is endopeptidase o-sialoglycoprotein, which cleaves at the glycoproteins of erythrocytes and can cause lysis. The other gene codes for superoxide dismutase (SOD), which may help detoxify reactive oxygen species, which protects ''M. haemofelis'' from oxidant damage when it is attached to the red blood cells of the host. In their research, it is stated that the reduction in the erythrocyte functions results in a short live span of the cells, which then can cause anemia for the host (5). | |||
In another study, the sequenced genome of the Langford 1 strain of ''M. haemofelis'' showed that this strain is capable of resulting in hemolytic anemia in specific-pathogen-free-derived felines. This was the first hemotropic mycoplasma species to be sequenced and annotated completely. This single, circular genome was 1,147,259 bp and had a GC content of 38.9%. The inspection of the genome revealed that glucose was the primary substrate for glycolysis of the bacteria. In addition, there appeared to be a different mechanism from other mycoplasmas for purine and pyrimidine metabolism because pentose phosphate pathways were unapparent. Furthermore, there were 31 tRNAs for the amino acids as well as three ribosomal operons. This research can develop on even further and soon answer the question of why this pathogen is complex and identify other mechanisms for how it evades the host immune system (1). | |||
==References== | ==References== | ||
[ | |||
1. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3133020/?tool=pubmed Barker, E.N. et al. (2011). Complete genome sequence of Mycoplasma haemofelis, a Hemotropic Mycoplasma. J. Bacteriol, 193.8, 2060-2061. ] | |||
2. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC155732/ Berent, L.M., and Messick, J.B. (2003). Physical map and genome sequencing survey of Mycoplasma haemofeli (Haemobartonella felis). American Society of Microbiology, 71.6, 3657-3662.] | |||
3. [http://jcm.asm.org/content/36/2/462.full.pdf+html Messick, J.B., Berent, L.M., and Cooper, S.K. (1998).Development and evaluation of PCR-Based assay for detection of Haemobartonella felis in cats and differentiation of H. felis from related bacteria by restriction fragment length polymorphism analysis. Journal of Clinical Microbiology, 36.2, 462-466.] | |||
4. [http://www.ncbi.nlm.nih.gov/sites/entrez?db=genome&cmd=Retrieve&dopt=Overview&list_uids=1663 Genome information for Mycoplasma haemofelis. NCBI Website. ] | |||
5. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3196708/ Santos et al. (2011). Genome of Mycoplasma haemofelis, unraveling its strategies for survival and persistence . Veterinary Research, 42.102, 1-16. ] | |||
6. [http://www.sciencedirect.com/science/article/pii/S0378113509003137 Tasker, S. et al. (2009). Description of outcomes of experimental infection with feline haemoplasmas: Copy numbers, haematology, Coomb’s testing and blood glucose concentrations. Vet. Microbiol., 139.3-4, 323-332. ] | |||
7. [http://www.sciencedirect.com/science/article/pii/S1098612X0190155X Tasker, S. and Lappin, MR. (2002). Haemobartonella felis: recent developments in diagnosis and treatment. Journal of Feline Medicine and Surgery, 4, 3-11.] | |||
8. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2880789/ Tasker, S. (2010). Haemotropic mycoplasmas: What’s their real significance in cats? J Feline Med Surg., 12.5, 369-381. ] | |||
9. [http://ijs.sgmjournals.org/content/35/4/497.full.pdf+html Tryon,V. and Pollack, D. (1985). Distinctions in Mollicutes Purine Metabolism: Pyrophosphate-Dependent Nucleoside Kinase and Dependence on Guanylate Salvage. International Journal of Systematic Bacteriology, 35.4, 497-501. ] | |||
10. [http://www.sciencedirect.com/science/article/pii/S0378113507003240 Willi, B. et al. (2007). From Haemobartonella to hemoplasma: Molecular methods provide new insights. Vet. Microbiology, 125.3-4, 197-209.] | |||
11. [http://www.vin.com/proceedings/Proceedings.plx?CID=WSAVA2010&PID=56139&Print=1&O=Generic Willi, B. et al. (2010). Haemotrophic mycoplasmas of cats and dogs: transmission, diagnosis, prevalence and importance in Europe. Schweiz Arch Tierheilkd, 152.5, 237-44.] | |||
Edited by Maggie Benson, students of [mailto:glim@rmc.edu Grace Lim-Fong] | Edited by Maggie Benson, students of [mailto:glim@rmc.edu Grace Lim-Fong] | ||
Latest revision as of 16:14, 5 December 2011
Characteristics of the symbiont/pathogen
Haemobartonella felis has been recently renamed to Mycoplasma haemofelis due to a closer relation to the genus Mycoplasma rather than the genus Bartonella (10). The genome of M. haemofelis has been sequenced, and in fact is the first hemotrophic mycoplasma species to be completely sequenced. In a study that compared two strains of the microbe, Ohio2 and Langford 1, they found that the complete genome of the circular chromosome was 1,155,937 base pairs. Even though the genome of M. haemofelis is small like other mycoplasmas, it still contains a large number of unique proteins (Figure 2). This large set of proteins is most likely due to its target for red blood cells, which may require a specific set of genes (5).
The phylogenetic classification of M. haemofelis is (4):
- Domain: Bacteria
- Phylum: Tenericutes
- Class: Mollicutes
- Family: Mycoplasmataceae
- Genus: Mycoplasma
- Species: Haemofelis
The cell morphology of M. haemofelis is a short coccus- or rod-shaped , gram negative bacteria. It also lacks a cell wall. Because the microbe is unable to be cultured,a microscopic identification from a blood smear is needed for diagnosis (3).
Characteristics of the host
The hosts for Mycoplasma haemofelis are felines. It is unclear of what breeds of felines are most commonly effected. However, it is found that felines of older non-pedigree breeds are more at risk for the infection that M. haemofelis even though younger felines can be affected (9). In other studies, they found that the infection is more common in felines from regions like Switzerland and other European countries (11).
Host-Symbiont Interaction
The interaction of the host, a cat, and the symbiont, Mycoplasma haemofelis, is one of a parasitic relationship. If the host acquires a relationship with the symbiont, the host will become anemic, which affects the host's red blood cell count. It is unclear of how the symbiont and disease is transmitted. However, some studies suggest that it is transmitted by fleas, so flea control is suggested for felines. Because it is known that the symbiont causes problems in the host's blood, it can be determined that another source of transmission is by blood, such as from blood donor cats that are in areas where the infection occurs (7). One of the symptoms for this disease are pale gums (Figure 3).
Since transmission is unclear, treatments are possible but not known to completely treat anemia for the infected feline. The most commonly used treatment is doxycycline that can be used for 14-21 days with one dosage a day. If the anemic conditions of the host are severe, a blood transfusion may be recommended (7).
The interaction is obligate, because the microbe M. haemofelis needs the host in order to carry out specific cellular functions. The class of M. haemofelis, Mollicutes, have been studied and determined to be dependent on hosts or other bacterium for guanine phosphoribosyl-transferase activity in order to carry out purine metabolism (9).
Molecular Insights into the Symbiosis
In some recent studies, the species Mycoplasma haemofelis has been compared to two other haemoplasma species, Candidatus M. haemominutum and Candidatus M. turicensis. In comparison of effects on the host of the symbiont, M. haemofelis causes haemolysis and C. M. haemominutum does not usually cause anemia. Coombs' tests and blood glucose concentration tests were used to determine the differences between these three species. Coomb's testing determines the presence for erythrocyte-bound antibodies and if the red blood cells become destroyed. The species M. haemofelis was the only species that tested positive for Coomb's test. In addition, the species C. M. turnicensis had lower glucose concentrations than the other two species as well as lower blood copy numbers (Figure 4). In conclusion of this study, they found that the species M. haemofelis was the only to develop macrocytic anemia (6).
Many genes of M. haemofelis have been studied. The two genes IMP (inosine-5′-monophosphate) dehydrogenase and GMP (guanosine 3′,5′-monophosphate) synthase have been found to encode proteins and partake in purine biosynthesis. This is significant because these enzymatic activities of these genes have not been found in Mollicutes. This raises the question if M. haemofelis needs a host in order to perform guanine biosynthesis,because it lacks the cellular components and functions for the biosynthesis. However, there is not enough research completed in order to determine this. Also, it is not clear what are the enzymes activities. In addition, the gene superoxide dismutase (SOD) is very unique to M. haemofelis because it is not found or sequenced in other mycoplasmas. However, there is no clear evidence of the genes' activity. Furthermore, it is found that this specie has many unique proteins and genes. Because of the unique genes, it is understood that this species of mycoplasma has adapted it self to colonize the red blood cell rather than mucosal surfaces like most mycoplasmas do and this is why it survives in its host (2).
Ecological and Evolutionary Aspects
In the past decade or so, Mycoplasma haemofelis has been difficult to diagnosis. However, with more recent research it is possible to diagnose for the infection by the polymerase chain reaction (PCR) rather than the older method of studying the red blood cells, which resulted in false positive and false negative due to other factors. These new PCR methods have contributed to the identification of two strains of M. haemofelis, Ohio strain and California strain. The Ohio strain causes a more severe infection than California strain and it could result in haemolytic anemia that could become fatal. The fact that strains are from different geological regions and that most felines affected are found in Europe may suggest that environmental factors play a role in regulating the symbiosis. However, M. haemofelis is still in the early stages of research because of its difficulty in culturing, so as of now environmental factors that regulate the parthenogenesis are not determined (7).
Recent Discoveries
In one study, the Mycoplasma haemofelis strain Ohio2 was used for genome sequencing in order to understand why the symbiont needs the host for metabolic pathways and resources in order to survive. M. haemofelis relies on its host because it lacks necessities for some metabolisms, such as pyrimidines and amino acids. However, this study represents some of the differences that M. haemofelis uses in some other metabolisms. For example, mycoplasmas have been thought to be incapable of fatty acid biosynthesis from acetyl-CoA because of a lack of genetic material. Yet, this study finds that M. haemofelis could be able to use glycerol in order to synthesize phospholipids. In addition, this study identified two virulence genes for M. haemofelis. One of these is endopeptidase o-sialoglycoprotein, which cleaves at the glycoproteins of erythrocytes and can cause lysis. The other gene codes for superoxide dismutase (SOD), which may help detoxify reactive oxygen species, which protects M. haemofelis from oxidant damage when it is attached to the red blood cells of the host. In their research, it is stated that the reduction in the erythrocyte functions results in a short live span of the cells, which then can cause anemia for the host (5).
In another study, the sequenced genome of the Langford 1 strain of M. haemofelis showed that this strain is capable of resulting in hemolytic anemia in specific-pathogen-free-derived felines. This was the first hemotropic mycoplasma species to be sequenced and annotated completely. This single, circular genome was 1,147,259 bp and had a GC content of 38.9%. The inspection of the genome revealed that glucose was the primary substrate for glycolysis of the bacteria. In addition, there appeared to be a different mechanism from other mycoplasmas for purine and pyrimidine metabolism because pentose phosphate pathways were unapparent. Furthermore, there were 31 tRNAs for the amino acids as well as three ribosomal operons. This research can develop on even further and soon answer the question of why this pathogen is complex and identify other mechanisms for how it evades the host immune system (1).
References
4. Genome information for Mycoplasma haemofelis. NCBI Website.
Edited by Maggie Benson, students of Grace Lim-Fong
This template is just a general guideline of how to design your site. You are not restricted to this format, so feel free to make changes to the headings and subheadings and to add or remove sections as appropriate.
