Providencia alcalifaciens: Difference between revisions
| Line 46: | Line 46: | ||
==Pathology== | ==Pathology== | ||
Infection symptoms include diarrhea, abdominal pain, and fever. ''P. alcalifaciens'' is believed to be involved in gastroenteritis. This bacteria is associated with travelers overseas. A problem of treating the infection from ''P. alcalifaciens'' is arising due to antibiotic resistance. | Infection symptoms include diarrhea, abdominal pain, and fever. ''P. alcalifaciens'' is believed to be involved in gastroenteritis. This bacteria is associated with travelers overseas. A problem of treating the infection from ''P. alcalifaciens'' is arising due to antibiotic resistance. There are two modes of entry that have been associated with ''P. alcalifaciens''. The bacteria is associated with actin condensation and endocytosis by a process of polymerizing the cytoskeletal components. ''P. alcalifaciens'' is also associated with interfering with tight junctions and entering into the intercellular spaces and thriving there. | ||
==References== | ==References== | ||
Revision as of 17:12, 9 May 2012
A Microbial Biorealm page on the genus Providencia alcalifaciens
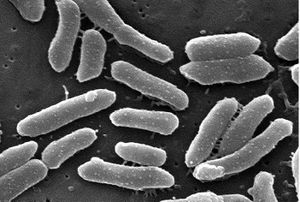
Classification
Higher order taxa
Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae; Providencia
Species
Providencia alcalifaciens
|
NCBI: [2] |
Description and significance
P. alcalifaciens is a rod-shaped gram-negative bacterium. This bacterium is associated with diarrhea in children and travelers. It is found in the gastrointestinal tract. Scientists have found that some strains of P. alcalifaciens are invasive to intestinal mucosa and other cell types in eukaryotes. In a study on the invasiveness of P. alcalifaciens on HEp-2 cells, the number of HEp-2 cells decreased within 6 hours after infection. This microbe causes actin condensation in the invaded cells. P. alcalifaciens thrives in environments around 37 degrees celsius especially for invasion. A study found that the microfilament inhibitor cytochalasin D inhibits the invasion by P. alcalifaciens.
Genome structure
|
NCBI: [3] |
According to the NCBI database, P. alcalifaciens is made up of 4,022 protein sequences encoded in a 4.03 Mb genome. The GC content is 41.8% 4. The genome was sequenced in 2008, isolated from human feces. Most P. alcalifaciens strains are organized in plasmids. Scientists are trying to determine whether the invasiveness and genome structure of the different bacterial strains have certain correlations concerning the presence of plasmids.
Cell and colony structure
Metabolism
Ecology
P. alcalifaciens is found in many different human and animal reservoirs. It is usually found in the gut or digestive tract and mostly seen in travelers who have traveled to developing countries. Young children and the elderly are at a higher risk for this infection.
Pathology
Infection symptoms include diarrhea, abdominal pain, and fever. P. alcalifaciens is believed to be involved in gastroenteritis. This bacteria is associated with travelers overseas. A problem of treating the infection from P. alcalifaciens is arising due to antibiotic resistance. There are two modes of entry that have been associated with P. alcalifaciens. The bacteria is associated with actin condensation and endocytosis by a process of polymerizing the cytoskeletal components. P. alcalifaciens is also associated with interfering with tight junctions and entering into the intercellular spaces and thriving there.
References
(1) Albert, M.J, M. Ansaruzzaman, et al. Characteristics of invasion by HEp-2 cells by Providencia alcalifaciens. J Med Microbiol. 1995 March; 42 [doi: 10.1099/00222615-42-3-186].
(2) Penner, J.L, M.A Preston. Difference among Providencia species in their in vitro susceptibilities to five antibiotics. Antimicrob Agents and Chemother. 1980 December; 18(6): 868-871.
(3) Sobreira, Marise, Nilma Leal, et al. Molecular analysis of clinical isolates of Providencia alcalifaciens. J Med Microbiol. 2001 Jan; 50(1) 29-34.
(4) http://www.ncbi.nlm.nih.gov/genome/?term=txid126385
Edited by Victoria Hughes of Dr. Lisa R. Moore, University of Southern Maine, Department of Biological Sciences, http://www.usm.maine.edu/bio
