Bifidobacterium choerinum: Difference between revisions
No edit summary |
|||
| Line 94: | Line 94: | ||
8) Scardovi V., Trovatelli L.D., Biavati B., Zani G. (1979). Bifidobacterium cuniculi, Bifidobacterium choerinum, Bifidobacterium boum, and Bifidobacterium pseudocatenulatum: four new species and their deoxyribonucieic acid homology relationships. Int. J. Syst. Bacteriol., 29: 291-311. | 8) Scardovi V., Trovatelli L.D., Biavati B., Zani G. (1979). Bifidobacterium cuniculi, Bifidobacterium choerinum, Bifidobacterium boum, and Bifidobacterium pseudocatenulatum: four new species and their deoxyribonucieic acid homology relationships. Int. J. Syst. Bacteriol., 29: 291-311. | ||
9) Miyake,T., Watanabe,K., Watanabe,T. and Oyaizu,H. <B>Phylogenetic Analysis of the Genus <I>Bifidobacterium</I> and Related Genera Based on 16S rDNA Sequences.</B> Microbiol. Immunol. 42 (10), 661-667 (1998) http://www.ncbi.nlm.nih.gov/nuccore/NR_037116.1 | |||
Revision as of 22:29, 29 April 2013
A Microbial Biorealm page on Bifidobacterium choerinum
Classification
Higher Order Taxa
Bacteria; Actinobacteria; Actinobacteridae; Bifidobacteriales; Bifidobacteriaceae; Bifidobacterium choerinum
Species
Species of the Genus Bifidobacterium include: B. adolescentis;B. angulatum;B. animalis; B. asteroids; B. bifidum; B. boum;B. breve;B. catenulatum; B. choerinum; B. coryneforme;B. cuniculi;B. denticolens; B. dentium; B. gallicum; B. gallinarum; B. indicum;B. infantis;B. inopinatum;B. lactis;B. longum;B. magnum;B. merycicum;B. minimum;B. pseudocatenulatum;B. pseudolongum;B. pullorum;B. ruminantium; B. saeculare; B. subtile; B. suis;B. thermacidophilum;B. thermophilum.
Description and Significance
The type strain is SUB06 from the Collection of the Istituto di Microbiologia Agraria, Universiti di Bologna, Italy. A culture of this strain has been deposited in the American Type Culture Collection under the number 27686. (8)
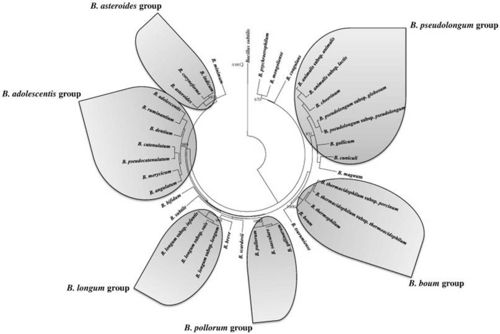
Members of the genus Bifidobacterium are some of the most common organisms in the human intestinal tract. (5) Bifidobacterium choerinum is part of the B. pseudolongum group which is a branch of the Actinobacteria. Bifidobacterium bacteria are classified as anaerobes that are known to be beneficial to human health. This group is expected to prove highly valuable for use in milk products, health promotion, and treatment of intestinal disorders. However, their sensitivity to O2 limits probiotic activity to solely anaerobic habitats. Recent research has reported that the Bifidobacterium strains exhibit various types of oxic growth. Low concentrations of O2 and CO2 can have a stimulatory effect on the growth of Bifidobacterium strains. (6)
Bifidobacterium choerinum is an autochthonous bifidobacterium species of the pig that is well adapted to the gut of pre-weaned piglets and shows potential probiotic properties. (2)
When researched it was shown that the DNA homology relationship between the other species of Bifidobacterium had the most relatedness to B. globosum and B. pseudolongum.(8)
Amino acid composition of the cell wall peptidoglycan: The interpeptide bridges of the cell wall mureins of the known strains are as follow: B. choerinum: Orn(Lys)-Ser-Ala2. (8)
Biochemical Reactions: Catalase is not produced. Pseudocatalase is not produced. Nitrate is not reduced. Indole is not produced. Gelatin is not liquefied. Skim milk is acidified and coagulated. Ammonia is not produced from urea or arginine. (8)
Genome Structure
Bifidobacterium chromosomes are circular shape with a high G-C content of 66.3 + or - 0.15mol%.(8)
The particular species of Bifidobacterium can be easily detected by using the species-specific PCR method for identifying Bifidobacterium strains isolated from human feces. (4)
The DNA of this species is essentially unrelated to that of any other species or homology group of Bifidobacterium (homologies range from 10 to 30%); exceptions are the DNAs of B. pseudolongum, B. globosum, and B. asteroides for which the homologies range from 25 to 60%. (8)
This topic has limited information found due to the little amount of research done with very few actual pictures of the species.
Cell and Colony Structure
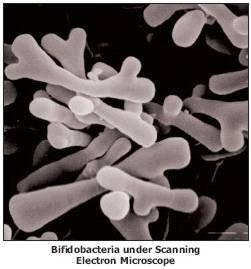
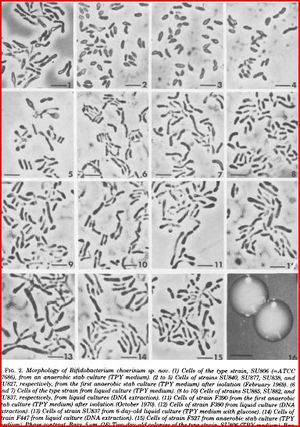
Bifidobacteria choerinum is a Gram-positive, anaerobic, branched rod-shaped bacterium. This particular type of bacteria are non-sporing and also non-motile. It is 1.5-3.5μm when freshly isolated but also appearing 10-12 μm when grown in a liquid TPY medium and has a V or Y-like appearance. The optimal temperature for growth is 37-41°C, no growth occurs below 20°C and above 46°C. When grown on a TPY medium, colonies appear smooth, convex, cream to white, glistening and soft. (8)
Bifidobacterium are acid-tolerant microorganisms, the optimal pH is between 6.5-7.3 with no growth occurring below 4 and above 8. (7) This bacterium has a hydrophobic character of bifidobacterium surface. (3) Bifidobacterium accumulate iron when it is presented in the ferrous oxidation state. This form is expected under anaerobic conditions like the human colon or cattle rumen. (3)
Metabolism
Bifidobacterium have been known to synthesize vitamins. It is an anaerobic microbe best grown in a TSA slant at 37°C. Significant amounts of polysaccharides normally galactose and glucose often associated with rhamnose. This bacterium is not considered to reduce nitrate. (3) It also will not develop in the presence of CO2 due to the effect of CO2 being non-detectable.
This type of bacterium ferments glucose via the phosphoketolase pathway and not via glycolysis. The typical fermentation end products are acetate and lactate. B. choerinum strains do not possess a fermentation pattern that differentiates them from other organisms. (8)
When residing in the intestines, they are able to ferment sugars to produce lactic acid. B. choerinum can ferment ribose, glucose, galactose, sucrose, maltose, melibiose, starch, and dextrin. It also showed weak and occasional fermentation of ribose. Mannose, trehalose, cellobiose, melezitose, mannitol, sorbitol, gluconate, salicin, glycerol, rhamnose or lactate are not fermented. (8)
It has also been shown to be resistant to aminoglycoside antibiotics while being susceptible to Beta-lactams. (3)
Ecology
This bacterium is ubiquitous, endosymbiotic inhabitant.
Pathology
No known pathology.
References
1) Euzeby, J.P. List of Prokaryotic Names with Standing in Nomenclature- Genus Bifidobacterium. 12 March 2013. http://www.bacterio.cict.fr/b/bifidobacterium.html
2) Maxwell FJ., Duncan SH., Hold G., Stewart CS. Isolation, Growth on Prebiotics and Probiotic Potential of Novel Bifidobacteria from Pigs. Anaerobe. (2004) Online.
3) Whitman, William B., Goodfellow, Micahel., Kampfer, Peter., et. (2012) Bergey's Manual of Systematic Bacteriology. Vol. 5.
4) Matsuki, Takahiro., Watanabe, Koichi., Fukuda, Masafumi., and Oyaizu, Hiroshi. (1999) Distribution of Bifidobacterial Species in Human Intestinal Microflora Examined with 16S rRNA-Gene-Targeted Species-Specific Primers Applied and Environmental Microbiology. 65:10, 4506-4512; 25 April 2013. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC91600/
5) Scardovi V. (1986). Genus Bifidobacterium Orla-Jensen 1924, 472al.. In: Sneath P.H.A., Mair N.S., Sharpe M.E., Holt J.G., eds, Bergey’s Manual of Systematic Bacteriology, Vol. 2, Williams and Wilkins, Baltimore, pp. 1418-1434.
6) Kawasaki, Shinji. (2011) Response of Bifidobacterium Species to Oxygen. Lactic Acid Bacteria and Bifidobacteria: Current Progress in Advanced Research. 25 April 2013. http://www.horizonpress.com/blogger/2011/03/response-of-bifidobacterium-species-to-oxygen.html
7) Biavati, B., Vescovo, M., Torriani, S., Bottazzi, V. Bifidobacteria: History, Ecology, Physiology and Applications. http://www.annmicro.unimi.it/full/50/biavati_50_117.pdf
8) Scardovi V., Trovatelli L.D., Biavati B., Zani G. (1979). Bifidobacterium cuniculi, Bifidobacterium choerinum, Bifidobacterium boum, and Bifidobacterium pseudocatenulatum: four new species and their deoxyribonucieic acid homology relationships. Int. J. Syst. Bacteriol., 29: 291-311.
9) Miyake,T., Watanabe,K., Watanabe,T. and Oyaizu,H. Phylogenetic Analysis of the Genus Bifidobacterium and Related Genera Based on 16S rDNA Sequences. Microbiol. Immunol. 42 (10), 661-667 (1998) http://www.ncbi.nlm.nih.gov/nuccore/NR_037116.1