Methanopyrus kandleri: evolutionary significance: Difference between revisions
No edit summary |
No edit summary |
||
| (48 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
{{Curated}} | |||
by Kiri Staiger | by Kiri Staiger | ||
==Introduction== | ==Introduction== | ||
<i>Methanopyrus kandleri</i> is a species of hyperthermophilic, halotolerant, methanogenic archaeon, originally isolated from sediments associated with undersea hydrothermal vent fields. [1] Currently the only species in its genus, <i>M. kandleri</i> is thought to be part of a separate lineage from all other known methanogens, possibly a lineage that diverged extremely early on, close to the time that the kingdoms of life were themselves diverging. [1] Whether or not this is the case is unclear; <i>M. kandleri</i> ’s exact phylogenetic placement, and its significance, have been points of controversy since the microbe’s discovery. [2, 3] Either way, both its unique lineage and its extremophilic nature set <i>M. kandleri</i> apart from its fellows, and it has been the subject of much study and debate. | [[File:Mkandcells.jpeg|600px|thumb|right|Figure 1: Micrograph of M. kandleri cells published in the original paper that first described the species. (1) [http://au8dt3yy7l.search.serialssolutions.com/?ctx_ver=Z39.88-2004&ctx_enc=info%3Aofi%2Fenc%3AUTF-8&rfr_id=info:sid/summon.serialssolutions.com&rft_val_fmt=info:ofi/fmt:kev:mtx:journal&rft.genre=article&rft.atitle=METHANOPYRUS-KANDLERI%2C+GEN+AND+SP-NOV+REPRESENTS+A+NOVEL+GROUP+OF+HYPERTHERMOPHILIC+METHANOGENS%2C+GROWING+AT+110-DEGREES-C&rft.jtitle=ARCHIVES+OF+MICROBIOLOGY&rft.au=KURR%2C+M&rft.au=HUBER%2C+R&rft.au=KONIG%2C+H&rft.au=JANNASCH%2C+HW&rft.date=1991-09-01&rft.pub=SPRINGER+VERLAG&rft.issn=0302-8933&rft.eissn=1432-072X&rft.volume=156&rft.issue=4&rft.spage=239&rft.epage=247&rft.externalDBID=n%2Fa&rft.externalDocID=A1991GF33100001¶mdict=en-US]]] | ||
<i>Methanopyrus kandleri</i> is a species of hyperthermophilic, halotolerant, methanogenic archaeon, originally isolated from sediments associated with undersea, hydrothermal, vent fields. [1] Currently the only species in its genus, <i>M. kandleri</i> is thought to be part of a separate lineage from all other known methanogens, possibly a lineage that diverged extremely early on, close to the time that the kingdoms of life were themselves diverging. [1] Whether or not this is the case is unclear; <i>M. kandleri</i> ’s exact phylogenetic placement, and its significance, have been points of controversy since the microbe’s discovery. [2, 3] Either way, both its unique lineage and its extremophilic nature set <i>M. kandleri</i> apart from its fellows, and it has been the subject of much study and debate. | |||
==Classification== | ==Classification== | ||
Assigning a classification to <i>M. kandleri</i> has been one of the main questions that researchers have debated about the microbe since its discovery in 1991. Although several different classifications have been suggested, the version currently accepted is: | Assigning a classification to <i>M. kandleri</i> has been one of the main questions that researchers have debated about the microbe since its discovery in 1991. Although several different classifications have been suggested, the version currently accepted is: | ||
Domain: Archaea | <br> | ||
Phylum: | <br>Domain: Archaea | ||
Class: Methanopyri | <br>Phylum: Euryarchaeota | ||
Order: Methanopyrales | <br>Class: Methanopyri | ||
Family: Methanopyraceae | <br>Order: Methanopyrales | ||
Genus: | <br>Family: Methanopyraceae | ||
<br>Genus: Methanopyrus | |||
<br>Species: kandleri | |||
==Species description== | |||
====Morphology==== | |||
[[File:Mkandbiofilm.jpeg|300px|thumb|right| Figure 2: Micrograph of <i>M. kandleri</i> cells (rods) growing in biofilm with <i>P furiosus</i> (cocci). <i>M. kandleri</i> is unusual in its ability to attach itself to glass under the shaking conditions in which this biofilm forms. Although the <i>furiosus</i> bacteria are unable to adhere to the smooth surface of the glass, they are quick to adhere to <i>M. kandleri</i>, resulting in a two species biofilm, with <i>kandleri</i> usually being layered over by several layers of <i>furiosus</i> cells. (4) [http://link.springer.com/article/10.1007%2Fs00203-008-0371-9/fulltext.html]]] | |||
<i>M. kandleri</i> presents as motile, rod-shaped cells, growing singly or in short chains of generally less than 10 individuals. Individual cells are typically between 8 and 10 microns in length, and half a micron in diameter. Occasional, very short, cells, only about .8 microns in length, are also observed, although whether these micro-cells are merely stunted or an example of intentional cell differentiation (as seen in heterocyst formation in cyanobacteria) is not currently known. Motility in <i>M. kandleri</i> is conferred by flagellar “tufts”, located at cell poles. Cells stain gram-positive; in addition to a psuedomurein inner envelope, cells possess the protein S-layer typical of archaeons. They also exhibit a slime-like layer that coats over the S-layer. It is thought that this slime layer is what allows <i>M. kandleri</i> to form the long, “raft-like” structures seen when the microbe is grown under shaking conditions. [1] It may also assist the microbe in adhering to surfaces. One experiment looking at archaeal bio-film formation found that <i>M. kandleri</i> was highly adherent, able to cling to even the smooth glass surface of a test tube under shaking conditions, where <i>Pyrococcus furiosus</i> (a different, hyperthermophilic, archaeon, whose preferred environmental conditions are comparable to those of <i>M. kandleri</i>) could not. [4] (Figure 2) Certainly, an enhanced ability to stick to one’s environment would not go amiss in <i>M. kandleri</i> ’s home in the walls of underwater thermal vents. | |||
<i>M. kandleri</i> | When cultured under less active conditions, <i>M. kandleri</i> may form irregularly shaped colonies, about 1 mm in diameter, that exhibit the blue-green morphology typical of methanogens. [1] | ||
<br><br><br> | |||
====Environmental Preferences==== | |||
Environmental Preferences | |||
The species was originally isolated from two sites—a “black smoker” hydrothermal vent site in the Guaymas Basin in the Gulf of California, and a shallower hydrothermal vent site at Kolbensey Ridge, off of Iceland. Although the samples were uniformly anaerobic, both sites otherwise exhibit incredible variation, in temperature—between 7° in the cooler sediments and 315° C in the vent-walls themselves—and pressure. The Guaymas Basin site is under 2000 meters of seawater; the Kolbensey Ridge site is much shallower at only around 100 m. [1] <i>M. kandleri</i> does not occupy the entire range of this incredibly diverse environment, but it does exhibit a fairly broad tolerance range, especially in regards to temperature and salinity. The optimum temperature for <i>M. kandleri</i> is 98° C, but the microbe can grow at temperatures as low 84°C and as high as 110°C—ten degrees above the boiling point of water. This extreme hyperthermophilism places <i>M. kandleri</i> among the most heat-tolerant organisms currently known. | The species was originally isolated from two sites—a “black smoker” hydrothermal vent site in the Guaymas Basin in the Gulf of California, and a shallower hydrothermal vent site at Kolbensey Ridge, off of Iceland. Although the samples were uniformly anaerobic, both sites otherwise exhibit incredible variation, in temperature—between 7° in the cooler sediments and 315° C in the vent-walls themselves—and pressure. The Guaymas Basin site is under 2000 meters of seawater; the Kolbensey Ridge site is much shallower at only around 100 m. [1] <i>M. kandleri</i> does not occupy the entire range of this incredibly diverse environment, but it does exhibit a fairly broad tolerance range, especially in regards to temperature and salinity. The optimum temperature for <i>M. kandleri</i> is 98° C, but the microbe can grow at temperatures as low 84°C and as high as 110°C—ten degrees above the boiling point of water. This extreme hyperthermophilism places <i>M. kandleri</i> among the most heat-tolerant organisms currently known. | ||
Salinity preference for <i>M. kandleri</i> is between 0.2 and 4%, with an optimal salinity of 2%. While not extreme, this does leave <i>M. kandleri</i> as a moderate halophile. The microbe has also been shown to tolerate high levels of <i>internal</i> salinity; like <i>Halobacteria</i> , <i>M. kandleri</i> builds up a high proportion of negatively charged molecules in its proteins and cell structures to counteract the positively charged salts. In the case of <i>M. kandleri</i> , the microbe may tolerate internal | Salinity preference for <i>M. kandleri</i> is between 0.2 and 4%, with an optimal salinity of 2%. While not extreme, this does leave <i>M. kandleri</i> as a moderate halophile. The microbe has also been shown to tolerate extremely high levels of <i>internal</i> salinity; like <i>Halobacteria</i> , <i>M. kandleri</i> builds up a high proportion of negatively charged molecules in its proteins and cell structures to counteract the positively charged salts. In the case of <i>M. kandleri</i>, the microbe may tolerate an internal concentration of potassium salts greater than 3M. [1,3] | ||
pH preference is near neutral, between 5.5 and 7; cells grow optimally at a pH of 6.5. | pH preference is near neutral, between 5.5 and 7; cells grow optimally at a pH of 6.5. | ||
Metabolism | ====Metabolism==== | ||
<i>M. kandleri</i> is an obligate, anaerobic, chemolithoautotroph; di-hydrogen gas and carbon dioxide are converted to methane, in a type of metabolism known as methanogenesis. This type of metabolism is found only in archaea, and plays an important role in the carbon cycle, especially in decomposition. Several different types of methanogenesis have been observed. The chemical equation below shows the reactants and involved in the variation of methanogenesis that <i>M. kandleri</i> utilizes. | <i>M. kandleri</i> is an obligate, anaerobic, chemolithoautotroph; di-hydrogen gas and carbon dioxide are converted to methane, in a type of metabolism known as methanogenesis. This type of metabolism is found only in archaea, and plays an important role in the carbon cycle, especially in decomposition. Several different types of methanogenesis have been observed. The chemical equation below shows the reactants and involved in the variation of methanogenesis that <i>M. kandleri</i> utilizes. | ||
| Line 40: | Line 45: | ||
Many methanogens may also use the carbon in organic compounds such as formate or methanol as an electron acceptor, but <i>M. kandleri</i> seems to strictly utilize carbon dioxide in this role. Hydrogen is similarly the sole electron donor—with one exception. Under normal circumstances, the presence of sulfur will only inhibit growth of <i>M. kandleri</i> ; however, sulfur is just similar enough to hydrogen that, under the right circumstances, such as during an exponential growth period where cells are growing and metabolizing rapidly, <i>M. kandleri</i> will mistake it for hydrogen and begin to use it as an electron donor. This is not unusual, per say; many microbes do in fact utilize sulfur in this way to no ill effect. However, one of the byproducts produced by this alternate chemical reaction is hydrogen sulfide (H2S), which appears to be toxic to the microbe, causing high rates of lysis. [1] | Many methanogens may also use the carbon in organic compounds such as formate or methanol as an electron acceptor, but <i>M. kandleri</i> seems to strictly utilize carbon dioxide in this role. Hydrogen is similarly the sole electron donor—with one exception. Under normal circumstances, the presence of sulfur will only inhibit growth of <i>M. kandleri</i> ; however, sulfur is just similar enough to hydrogen that, under the right circumstances, such as during an exponential growth period where cells are growing and metabolizing rapidly, <i>M. kandleri</i> will mistake it for hydrogen and begin to use it as an electron donor. This is not unusual, per say; many microbes do in fact utilize sulfur in this way to no ill effect. However, one of the byproducts produced by this alternate chemical reaction is hydrogen sulfide (H2S), which appears to be toxic to the microbe, causing high rates of lysis. [1] | ||
====Genome type==== | |||
<i>Methanopyrus kandleri</i> possesses a single, circular chromosome 1,694,969 base pairs in length, containing genes coding for 39 different structural RNAs, and 1,692 different proteins. Of these proteins, only a very few are involved in gene regulation or regulation signaling pathways; fewer than one would predict, even when one takes into consideration that hyperthermophiles as a group are generally less regulated than other microbes. <i>M. kandleri</i> ’s genome also encodes fewer DNA binding proteins, also associated with regulatory activity, than any other known archaeon; this suggests that regulatory activity in <i>M. kandleri</i> is highly reduced, and response pathways may be relatively few. [3] | <i>Methanopyrus kandleri</i> possesses a single, circular chromosome 1,694,969 base pairs in length, containing genes coding for 39 different structural RNAs, and 1,692 different proteins. Of these proteins, only a very few are involved in gene regulation or regulation signaling pathways; fewer than one would predict, even when one takes into consideration that hyperthermophiles as a group are generally less regulated than other microbes. <i>M. kandleri</i> ’s genome also encodes fewer DNA binding proteins, also associated with regulatory activity, than any other known archaeon; this suggests that regulatory activity in <i>M. kandleri</i> is highly reduced, and response pathways may be relatively few. [3] | ||
| Line 48: | Line 53: | ||
The G+C content is about 60%, which is unusually high compared to most other methanogens, which average only 33% G+C . [1] | The G+C content is about 60%, which is unusually high compared to most other methanogens, which average only 33% G+C . [1] | ||
Novel proteins and features: | ==Novel proteins and features== | ||
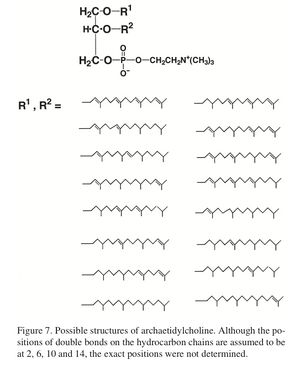
[[File:Lipidstructure.jpeg|300px|thumb|right| Figure 3: Molecular structure of a novel, choline containing, allyl ether-linked phospholipid isolated from <i>M. kandleri</i>, along with other novel lipids. archaetidylcholine (2,3-di- O-phytanyl-sn-glycerophosphocholine) Figure from Nishihara et al (13)[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2685562/pdf/Archaea-01-123.pdf] ]] | |||
A number of novel proteins and other compounds have been isolated from <i>M. kandleri</i> , and are of interest in perhaps shedding light on both its phylogenetic status and its highly thermophilic, halophilic existence. Many hyperthermophilic proteins and enzymes have been described from <i>M. kandleri</i> , and some have found applications in the world of biotechnology. Some of the more interesting discoveries, in terms of either evolutionary significance or possible applications, are outlined in this section. | A number of novel proteins and other compounds have been isolated from <i>M. kandleri</i> , and are of interest in perhaps shedding light on both its phylogenetic status and its highly thermophilic, halophilic existence. Many hyperthermophilic proteins and enzymes have been described from <i>M. kandleri</i> , and some have found applications in the world of biotechnology. Some of the more interesting discoveries, in terms of either evolutionary significance or possible applications, are outlined in this section. | ||
====Novel Psuedomurien Variate==== | |||
Many archaeons possess a cell wall consisting of pseudomurein. Also called pseudopeptidoglycan, pseudomurein acts much as peptidoglycan does in bacterial cell walls, forming a thick, multi-layered envelope around the cell. Ordinarily, pseudomurein is made up of N-acetylglucosamine and N-acetyltalosaminuronic acid. However, <i>Methanopyrus kandleri</i> substitutes L-orthinine where N-acetylglucosamine ordinarily would go. | Many archaeons possess a cell wall consisting of pseudomurein. Also called pseudopeptidoglycan, pseudomurein acts much as peptidoglycan does in bacterial cell walls, forming a thick, multi-layered envelope around the cell. Ordinarily, pseudomurein is made up of N-acetylglucosamine and N-acetyltalosaminuronic acid. However, <i>Methanopyrus kandleri</i> substitutes L-orthinine where N-acetylglucosamine ordinarily would go. | ||
====Novel Core Lipid==== | |||
A number of novel lipids have been isolated from <i>M. kandleri</i>. The figure at right shows the structure of one of these lipids, 2,3-di- O-phytanyl-sn-glycerophosphocholine (Fig. 3). In addition, the microbe was found to utilize a novel core lipid. This lipid, an unsaturated, ether-linked lipid identified as 2,3-di-O-geranylgeranyl-sn-glycerol, is of a type of lipids (terpenoid lipids) thought to be a ‘primitive feature’ of many microbes. The presence of this terpenoid lipid may, in combination with other evidence (as detailed later in this article), provide some evidence for the claim that <i>M. kandleri</i> is of an early diverging branch from the <i>Euryarchaeota</i> group. [12] | |||
====Novel DNA-binding proteins==== | |||
Sequence analysis of the <i>M. kandleri</i> genome shows a relative deficiency in this microbe of the helix-turn-helix and Arc-MetJ | Sequence analysis of the <i>M. kandleri</i> genome shows a relative deficiency in this microbe of the helix-turn-helix and Arc-MetJ type DNA binding proteins that are usually found in abundance across both archaeons and bacteria. Instead, <i>M. kandleri</i> appears to utilize (at least) 3 novel DNA binding proteins. All are relatively small molecules, with molecular weights of 7, 10, and 30 kDa. The structure of the 7 kDa binding protein has been determined to be dimeric, consisting of a larger alpha-helical protein associated with a smaller, N-terminal beta-strand. Structural properties, as well as apparent regulatory similarities, suggest that this binding protein may be a relative of the ribbon-helix-helix family of transcription factors that are involved in DNA binding and gene regulation in bacteria, notably <i>Pseudomonas</i> bacteria. [10] | ||
====Hetero-dimeric reverse DNA gyrase==== | |||
Reverse DNA gyrases, which positively supercoil DNA when activated by ATP, have been found to be common across nearly all hyperthermophiles, both archaeons and bacteria. However, the <i>M. kandleri</i> DNA reverse gyrase is the first hetero-dimeric DNA reverse gyrase to be described. The protein consists of two sub-units—a larger (138 kDa) subunit that associates with ATP and hydrolyses it to release energy; and a smaller (43 kDa) sub-unit that binds covalently to DNA as the protein works to create supercoils. | Reverse DNA gyrases, which positively supercoil DNA when activated by ATP, have been found to be common across nearly all hyperthermophiles, both archaeons and bacteria. However, the <i>M. kandleri</i> DNA reverse gyrase is the first hetero-dimeric DNA reverse gyrase to be described. The protein consists of two sub-units—a larger (138 kDa) subunit that associates with ATP and hydrolyses it to release energy; and a smaller (43 kDa) sub-unit that binds covalently to DNA as the protein works to create supercoils. | ||
The presence of these two sub-units within the gyrase, as opposed to having a single unit protein, is thought to help stabilize the protein at the incredibly high temperatures <i>M. kandleri</i> is subject to. In addition, high temperatures appear to be required for the two sub-units to form the stable heterodimer observed. [9] | The presence of these two sub-units within the gyrase, as opposed to having a single unit protein, is thought to help stabilize the protein at the incredibly high temperatures <i>M. kandleri</i> is subject to. In addition, high temperatures appear to be required for the two sub-units to form the stable heterodimer observed. [9] | ||
Novel and modified RNA nucleosides | ====Novel and modified RNA nucleosides==== | ||
<i>M. kandleri</i> shows an unusual and highly diverse pattern of post-transcriptional modifications to tRNA nucleosides—4 entirely novel nucleosides have been isolated from this microbe. Modifications to these nucleosides effect structure, behavior, and stability of the tRNA strands each helps to form. The high degree of modification seen in <i>M. kandleri</i> may have to do with the extreme nature of its environment and the high demands that environment places on its proteins in terms of maintaining stability. [11] | <i>M. kandleri</i> shows an unusual and highly diverse pattern of post-transcriptional modifications to tRNA nucleosides—4 entirely novel nucleosides have been isolated from this microbe. Modifications to these nucleosides effect structure, behavior, and stability of the tRNA strands each helps to form. The high degree of modification seen in <i>M. kandleri</i> may have to do with the extreme nature of its environment and the high demands that environment places on its proteins in terms of maintaining stability. [11] | ||
Questions and Controversy- | ==Questions and Controversy- Placing <i>M. kandleri</i> within the evolutionary tree of life== | ||
Since its discovery, the placement of <i>M. kandleri</i> within a broader phylogenetic context has been a highly contested avenue of research. At different times, it has been thought to be many different things—a representative of a new class within <i>Euryarchaeota</i> , only very distantly related to its fellow methanogens [1, 2]; alternatively, quite closely related to other methanogens [3]; or, even more alternatively, quite unrelated from just about everything, and possibly a link to finding the much lauded Last Universal Common Ancestor [5]. Which one of these <i>M. kandleri</i> actually is, is still up to debate. | Since its discovery, the placement of <i>M. kandleri</i> within a broader phylogenetic context has been a highly contested avenue of research. At different times, it has been thought to be many different things—a representative of a new class within <i>Euryarchaeota</i> , only very distantly related to its fellow methanogens [1, 2]; alternatively, quite closely related to other methanogens [3]; or, even more alternatively, quite unrelated from just about everything, and possibly a link to finding the much lauded Last Universal Common Ancestor [5]. Which one of these <i>M. kandleri</i> actually is, is still up to debate. | ||
====Initial Classification==== | |||
The researchers who initially isolated the microbe placed it in a separate, deeper-branching, clade from other methanogens based on the following observations: | The researchers who initially isolated the microbe placed it in a separate, deeper-branching, clade from other methanogens based on the following observations: | ||
First, the G+C content of <i>Methanopyrus</i> was much higher than that of other known methanogens. At 60%G+C, <i>Methanopyrus</i> has almost twice the G+C in its genome as other methanogens. Second, the amount of homologous DNA sequences observed between <i>Methanopyrus</i> and other methanogens was, in their words, “insignificant.” Third, lipid analysis of the structures of <i>Methanopyrus</i> revealed a very different set of core lipids than what ought to be a closely related archaeons, <i>Methanococcus</i> . Where <i>Methanococcus</i> ’ core lipids are 85% comprised of cyclic dibisphytanyl tetraethers, <i>Methanopyrus</i> ’ core lipid was 2,3-di-O-phytanyl-glycerol, a very different type of lipid. Fourth, <i>Methanopyrus</i> had a higher halo-tolerance than other <i>Methanobacteriales</i> species. Fifth, <i>Methanopyrus</i> exhibited a novel type of psuedomurein in its cell wall, a variant that substituted L-ornithine for N-acetylglucosamine, and had not been observed previously. Sixth, and perhaps most convincingly, phylogenetic analysis of the microbe’s 16s RNA sequence placed it quite distantly from all known methanogens. In fact, 16s RNA analysis placed it so very distantly from other methanogens that the researchers designated it in a new class—Class Methanopyri, which they had branching off from the Phylum <i>Euryarchaeota</i> significantly earlier than any other known branch. [1] | First, the G+C content of <i>Methanopyrus</i> was much higher than that of other known methanogens. At 60%G+C, <i>Methanopyrus</i> has almost twice the G+C in its genome as other methanogens. Second, the amount of homologous DNA sequences observed between <i>Methanopyrus</i> and other methanogens was, in their words, “insignificant.” Third, lipid analysis of the structures of <i>Methanopyrus</i> revealed a very different set of core lipids than what ought to be a closely related archaeons, <i>Methanococcus</i> . Where <i>Methanococcus</i> ’ core lipids are 85% comprised of cyclic dibisphytanyl tetraethers, <i>Methanopyrus</i> ’ core lipid was 2,3-di-O-phytanyl-glycerol, a very different type of lipid. Fourth, <i>Methanopyrus</i> had a higher halo-tolerance than other <i>Methanobacteriales</i> species. Fifth, <i>Methanopyrus</i> exhibited a novel type of psuedomurein in its cell wall, a variant that substituted L-ornithine for N-acetylglucosamine, and had not been observed previously. Sixth, and perhaps most convincingly, phylogenetic analysis of the microbe’s 16s RNA sequence placed it quite distantly from all known methanogens. In fact, 16s RNA analysis placed it so very distantly from other methanogens that the researchers designated it in a new class—Class Methanopyri, which they had branching off from the Phylum <i>Euryarchaeota</i> significantly earlier than any other known branch. [1] | ||
====Support for a separate lineage==== | |||
[[File:Trees2.jpeg|400px|thumb|right| Figure 4: Proposed phylogenies that would place <i>M. kandleri</i> in a separately diverging group from the other methanogens, and possibly closer to the <i>Eocyte</i> lineage. (2) [http://ijs.sgmjournals.org/content/46/1/348.short] ]] | |||
In the research since <i>Methanopyrus</i> ’ initial discovery in 1991, there has been some support for this theory. Analysis of <i>M. kandleri</i> ’s 16s RNA sequence suggests that, although a Euryarchaote, <i>Methanopyrus</i> shares a higher degree of relatedness to the chrenarchaeota group that seen in any other <i>Euryarchaeote</i> , which in turn suggests that <i>M. kandleri</i> may have diverged very early on. A 1996 study used a Jukes-Cantor distance projection to approach the problem qualitatively, looking at distances between 16s RNA sequences and regions of the 1-α elongation factor for <i>M. kandleri</i> and proposed relations, including methanogens. The tree generated by this projection again placed <i>Methanopyrus</i> at a deep branch within <i>Euryarchaeota</i> , existing either within or proximal to archaebacteria. [2] (Figure 4) A similar study using the sequences from an operon common across methanogens, coding for methyl coenzyme M reductase, also supported the conclusion that <i>Methanopyrus</i> stands separate from other methanogens. [6] | |||
If this view of <i>Methanopyrus</i> phylogeny is correct and <i>Methanopyrus</i> is of a separate lineage than other methanogens, this would indicate that methanogenesis as a metabolism has arisen at least two separate times. Perhaps more importantly, because many phylogenetic analyses of <i>Methanopyrus</i> place it as a representative of a very deep branching group, some have looked to <i>Methanopyrus</i> as perhaps the most proximal known relative of the Last Universal Common Ancestor, or LUCA. Although previous efforts to locate the LUCA placed it somewhere in the bacterial realm, more recent analyses, incorporating a greater diversity of organisms, especially the previously under-represented archaeons, tell a different story. Phylogenetic distances based on tRNA sequences, which show much lower rates of horizontal transfer than protein coding genes and, as such, are more likely to generate accurate phylogenetic trees over this kind of time-frame, suggest that the LUCA falls somewhere between <i>Euryarchaeota</i> and <i>Crenarchaeota</i> —just about, in fact, where <i>Methanopyrus kandleri</i> seems to fit. [8] If this is the case, than <i>M. kandleri</i> and its genome can tell us a lot about the early divergence of life. | |||
[ | ====Support for a monophyletic view of <i>M. kandleri</i> and the methanogens==== | ||
[[File:Trees1.jpeg|350px|thumb|right| Figure 5: Alternatively proposed phylogenies for <i>M. kandleri</i> ("Metka") based on COG-based DNA distance analysis. These phylogenies place <i>Methanopyrus</i> in a single, closely related clade with the other methanogens. (3) [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC123701/]]] | |||
As with any radical claim in science, the proposal of a <i>Methanopyrus</i>-proximal LUCA met with immediate refutal. Opposing this view—opposing, in fact, the entire idea that <i>Methanopyrus</i> is of a different lineage than its fellow methanogens—we have a second set of research. One study used full DNA sequence data and the idea of COGs—Clusters of Orthologous Groups, sets of gene orthologs that are conserved across different phylogenetic groups—to construct a very different phylogeny, that grouped <i>M. kandleri</i> in one lineage with other methanogens. According to this work, about 73% of the proteins coded for in <i>M. kandleri</i> ’s genome are part of COGs shared across a broad spectrum of life; many of the remaining proteins are common across methanogens, which supports the proposal of a single methanogenic lineage. In the trees generated by this COG analysis, <i>M. kandleri</i> is shown as most proximal to either <i>Methanococcus thermoautotrophicum</i> or <i>Methanococcus jannaschii</i>. | |||
The sequence data for these three species—<i>Methanococcus thermoautotrophicum</i>, <i>Methanococcus jannaschii</i> , and <i>Methanopyrus kandleri</i> —is somewhat ambiguous as to who exactly is most related to whom. All three share COGs that are common across methanogens. <i>M. jannaschii</i> and <i>M. thermoautotrophicum</i> share a partial duplication in a recently described operon. The placement and order of genes and operons is more similar between <i>M. thermoautotrophicum</i> and <i>M. kandleri</i> . Depending on what metric we pick, these three species arrange themselves into various relationships. But, despite this ongoing fine-tuning, this research makes a clear case for placing <i>Methanopyrus</i> within a single lineage with the methanogens. (Figure 5) [3] | |||
Further muddying the waters, it has been suggested that many of the phylogenetic analyses that have placed <i>M. kandleri</i> in a separate lineage may be reflective not of an ancient divergence but of an unusually high mutation rate. Hyperthermophilic conditions such as those that <i>M. kandleri</i> thrives in are known to effect the stability of DNA secondary structures, causing hyperthermophilic microorganisms to exhibit higher than normal rates of sequence mutation. So one school of thought would hold that this relative genetic instability would skew any genetic distance analyses of <i>M. kandleri</i> towards predicting a much earlier divergence than actually occurred. On the other hand, mechanisms have been observed in many non-archaeal hyperthermophiles that mitigate wholly or entirely this effect, lowering their mutation rates to something more comparable to non-hyperthermophiles. These mechanisms, or similar mechanisms, may be in effect in <i>M. kandleri</i>-- however, because the DNA repair genes that encode these mechanisms are not well known in archaea, we just don't know at this point whether this is the case. (14) As seems to be often the case with <i>M. kandleri</i>, we are left questioning. | |||
So—what to do with <i>Methanopyrus</i>, in the end? Is it a radically new lineage of methanogen, presenting new insight into the last universal origins of life itself? Is it nothing so bold, just another methanogen? With evidences that seem to point in all directions, this question is not yet resolved, and may not be any time soon. Which evidences do we choose? Do we base our phylogenetic analysis off of 16s RNA? Whole genome analysis? tRNA sequences? Whichever way we do it, we seem to get a different result. The paradox of <i>M. kandleri</i> remains. | |||
[8] | ==References== | ||
[http://au8dt3yy7l.search.serialssolutions.com/?ctx_ver=Z39.88-2004&ctx_enc=info%3Aofi%2Fenc%3AUTF-8&rfr_id=info:sid/summon.serialssolutions.com&rft_val_fmt=info:ofi/fmt:kev:mtx:journal&rft.genre=article&rft.atitle=METHANOPYRUS-KANDLERI%2C+GEN+AND+SP-NOV+REPRESENTS+A+NOVEL+GROUP+OF+HYPERTHERMOPHILIC+METHANOGENS%2C+GROWING+AT+110-DEGREES-C&rft.jtitle=ARCHIVES+OF+MICROBIOLOGY&rft.au=KURR%2C+M&rft.au=HUBER%2C+R&rft.au=KONIG%2C+H&rft.au=JANNASCH%2C+HW&rft.date=1991-09-01&rft.pub=SPRINGER+VERLAG&rft.issn=0302-8933&rft.eissn=1432-072X&rft.volume=156&rft.issue=4&rft.spage=239&rft.epage=247&rft.externalDBID=n%2Fa&rft.externalDocID=A1991GF33100001¶mdict=en-US] 1. Margit Kurr, Robert Huber, Helmut Konig, Holger W. Jannasch, Hans Fricke, Antonio Trineone, Jakob K. Kristjansson, and Karl O. Stetter. 1991. <i>Methanopyrus kandleri</i> ,gen. and sp. nov. represents a novel group of hyperthermophilic methanogens, growing at 110° C. Arch Microbiol 156:239—247. | |||
[ | [http://ijs.sgmjournals.org/content/46/1/348.short] 2. Rivera, M.C., and Lake, J. 1996. The Phylogeny of <i>Methanopyrus kandleri</i> . IJSEM 46: 348-351. | ||
[ | [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC123701/] 3. Slesarev AI, Mezhevaya KV, Makarova KS, Polushin NN, Shcherbinina OV, et al. 2002. The complete genome of hyperthermophile <i>Methanopyrus kandleri</i> AV19 and monophyly of archaeal methanogens. Proc Natl Acad Sci U S A 99: 4644–4649. | ||
[ | [http://link.springer.com/article/10.1007%2Fs00203-008-0371-9/fulltext.html] 4. Simone Schopf, Gerhard Wanner, Reinhard Rachel, and Reinhard Wirth. 2008. An archaeal bi-species biofilm formed by Pyrococcus furiosus and <i>Methanopyrus kandleri</i> . Archives of Microbiology 190: 371-377. | ||
[http://link.springer.com/article/10.1007%2Fs13213-010-0154-9] 5. Zhiliang Yu, and Shanxiang Xu. 2011. Search for a <i>Methanopyrus</i> -proximal last universal common ancestor based on comparative-genomic analysis. Annals of Microbiology 61: 397-401. | |||
[http://ijs.sgmjournals.org/content/46/4/1170.full.pdf] 6. JORK NOLLING, AMY ELFNER, JOHN R. PALMER, VANESSA J. STEIGERWALD, TODD D. PIHL, JAMES A. LAKE, AND JOHN N. REEVE. 1996. Phylogeny of Methanopyms kandleri Based on Methyl Coenzyme M Reductase Operons. INTERNATIONAL JOURNAL oF SYSTEMATIC BACTERIOLOGY. 46: 1170-1173. | |||
[http://link.springer.com/article/10.1007/s00239-009-9297-3] 7. Zhiliang Yu, Ken Takai, Alexei Slesarev, Hong Xue and J. Tze-Fei Wong. 2009. Search for Primitive <i>Methanopyrus</i> Based on Genetic Distance Between Val- and Ile-tRNA Synthetases. Journal of Molecular Evolution 69: 386-394 | |||
[http://www.sciencedirect.com/science/article/pii/S0723202011803085] 8. S. Burggraf, K.O. Stetter, P. Rouviere, C.R. Woese. 1991. <i>Methanopyrus kandleri</i> : An Archaeal Methanogen Unrelated to all Other Known Methanogens. Systematic and Applied Microbiology 14: 346-351. | |||
[ | [http://www.jbc.org/content/272/21/13986.abstract] 9. Regis Krah, Mary H. O’Dea and Martin Gellert. 1997. RECONSTITUTION OF AN ACTIVE EXTREMOZYME FROM ITS TWO RECOMBINANT SUBUNITS. The Journal of Biological Chemistry, 272, 13986-13990. | ||
[http://nar.oxfordjournals.org/content/30/3/685.full] 10. Nikolai A. Pavlov, Dmitry I. Cherny, Igor V. Nazimov, Alexei I. Slesarev, and Vinod Subramaniam. 2002. Identification, cloning and characterization of a new DNA-binding protein from the hyperthermophilic methanogen <i>Methanopyrus kandleri</i> . Nucl. Acids Res. (2002) 30 (3): 685-694. | |||
[http://www.febsletters.org/article/S0014-5793(05)00474-6/fulltext] 11. Sauerwald, Anselm1; Sitaramaiah, Devarasetty2; McCloskey, James A.2,3; Söll, Dieter1; Crain, Pamela F.. 2005. N6-Acetyladenosine: A new modified nucleoside from <i>Methanopyrus kandleri</i> tRNA 579: 2807-2810. | |||
[ | [http://www.sciencedirect.com/science/article/pii/S0723202011804637] 12. HAFENBRADL, D; KELLER, M; THIERICKE, R; STETTER, KO. 1993. A NOVEL UNSATURATED ARCHAEAL ETHER CORE LIPID FROM THE HYPERTHERMOPHILE <I>METHANOPYRUS</i> -KANDLERI. SYSTEMATIC AND APPLIED MICROBIOLOGY 16: 165-169. | ||
[ | [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2685562/pdf/Archaea-01-123.pdf] 13. MASATERU NISHIHARA, HIROYUKI MORII, KOJI MATSUNO, MAMI OHGA, KARL O. STETTER, and YOSUKE KOGA. 2002. Structural analysis by reductive cleavage with LiAlH4 of an allyl ether choline-phospholipid, archaetidylcholine, from the hyperthermophilic methanoarchaeon Methanopyrus kandleri. Archaea 1, 123-131. | ||
[ | [http://onlinelibrary.wiley.com/store/10.1046/j.1365-2958.1998.00853.x/asset/j.1365-2958.1998.00853.x.pdf;jsessionid=C522A13C34D16562B28F4EDF13C8E579.f02t04?v=1&t=huyhtbm6&s=fb2fab5950c66fcdd360cba54a1f4f6a42910a45] 14. Dennis W. Grogan. 1998. Hyperthermophiles and the problem of DNA instability. Molecular Microbiology 28(6):1043–1049. | ||
Latest revision as of 15:31, 2 October 2015
by Kiri Staiger
Introduction

Methanopyrus kandleri is a species of hyperthermophilic, halotolerant, methanogenic archaeon, originally isolated from sediments associated with undersea, hydrothermal, vent fields. [1] Currently the only species in its genus, M. kandleri is thought to be part of a separate lineage from all other known methanogens, possibly a lineage that diverged extremely early on, close to the time that the kingdoms of life were themselves diverging. [1] Whether or not this is the case is unclear; M. kandleri ’s exact phylogenetic placement, and its significance, have been points of controversy since the microbe’s discovery. [2, 3] Either way, both its unique lineage and its extremophilic nature set M. kandleri apart from its fellows, and it has been the subject of much study and debate.
Classification
Assigning a classification to M. kandleri has been one of the main questions that researchers have debated about the microbe since its discovery in 1991. Although several different classifications have been suggested, the version currently accepted is:
Domain: Archaea
Phylum: Euryarchaeota
Class: Methanopyri
Order: Methanopyrales
Family: Methanopyraceae
Genus: Methanopyrus
Species: kandleri
Species description
Morphology

M. kandleri presents as motile, rod-shaped cells, growing singly or in short chains of generally less than 10 individuals. Individual cells are typically between 8 and 10 microns in length, and half a micron in diameter. Occasional, very short, cells, only about .8 microns in length, are also observed, although whether these micro-cells are merely stunted or an example of intentional cell differentiation (as seen in heterocyst formation in cyanobacteria) is not currently known. Motility in M. kandleri is conferred by flagellar “tufts”, located at cell poles. Cells stain gram-positive; in addition to a psuedomurein inner envelope, cells possess the protein S-layer typical of archaeons. They also exhibit a slime-like layer that coats over the S-layer. It is thought that this slime layer is what allows M. kandleri to form the long, “raft-like” structures seen when the microbe is grown under shaking conditions. [1] It may also assist the microbe in adhering to surfaces. One experiment looking at archaeal bio-film formation found that M. kandleri was highly adherent, able to cling to even the smooth glass surface of a test tube under shaking conditions, where Pyrococcus furiosus (a different, hyperthermophilic, archaeon, whose preferred environmental conditions are comparable to those of M. kandleri) could not. [4] (Figure 2) Certainly, an enhanced ability to stick to one’s environment would not go amiss in M. kandleri ’s home in the walls of underwater thermal vents.
When cultured under less active conditions, M. kandleri may form irregularly shaped colonies, about 1 mm in diameter, that exhibit the blue-green morphology typical of methanogens. [1]
Environmental Preferences
The species was originally isolated from two sites—a “black smoker” hydrothermal vent site in the Guaymas Basin in the Gulf of California, and a shallower hydrothermal vent site at Kolbensey Ridge, off of Iceland. Although the samples were uniformly anaerobic, both sites otherwise exhibit incredible variation, in temperature—between 7° in the cooler sediments and 315° C in the vent-walls themselves—and pressure. The Guaymas Basin site is under 2000 meters of seawater; the Kolbensey Ridge site is much shallower at only around 100 m. [1] M. kandleri does not occupy the entire range of this incredibly diverse environment, but it does exhibit a fairly broad tolerance range, especially in regards to temperature and salinity. The optimum temperature for M. kandleri is 98° C, but the microbe can grow at temperatures as low 84°C and as high as 110°C—ten degrees above the boiling point of water. This extreme hyperthermophilism places M. kandleri among the most heat-tolerant organisms currently known.
Salinity preference for M. kandleri is between 0.2 and 4%, with an optimal salinity of 2%. While not extreme, this does leave M. kandleri as a moderate halophile. The microbe has also been shown to tolerate extremely high levels of internal salinity; like Halobacteria , M. kandleri builds up a high proportion of negatively charged molecules in its proteins and cell structures to counteract the positively charged salts. In the case of M. kandleri, the microbe may tolerate an internal concentration of potassium salts greater than 3M. [1,3]
pH preference is near neutral, between 5.5 and 7; cells grow optimally at a pH of 6.5.
Metabolism
M. kandleri is an obligate, anaerobic, chemolithoautotroph; di-hydrogen gas and carbon dioxide are converted to methane, in a type of metabolism known as methanogenesis. This type of metabolism is found only in archaea, and plays an important role in the carbon cycle, especially in decomposition. Several different types of methanogenesis have been observed. The chemical equation below shows the reactants and involved in the variation of methanogenesis that M. kandleri utilizes.
CO2 + 4 H2 → CH4 + 2H2O
Many methanogens may also use the carbon in organic compounds such as formate or methanol as an electron acceptor, but M. kandleri seems to strictly utilize carbon dioxide in this role. Hydrogen is similarly the sole electron donor—with one exception. Under normal circumstances, the presence of sulfur will only inhibit growth of M. kandleri ; however, sulfur is just similar enough to hydrogen that, under the right circumstances, such as during an exponential growth period where cells are growing and metabolizing rapidly, M. kandleri will mistake it for hydrogen and begin to use it as an electron donor. This is not unusual, per say; many microbes do in fact utilize sulfur in this way to no ill effect. However, one of the byproducts produced by this alternate chemical reaction is hydrogen sulfide (H2S), which appears to be toxic to the microbe, causing high rates of lysis. [1]
Genome type
Methanopyrus kandleri possesses a single, circular chromosome 1,694,969 base pairs in length, containing genes coding for 39 different structural RNAs, and 1,692 different proteins. Of these proteins, only a very few are involved in gene regulation or regulation signaling pathways; fewer than one would predict, even when one takes into consideration that hyperthermophiles as a group are generally less regulated than other microbes. M. kandleri ’s genome also encodes fewer DNA binding proteins, also associated with regulatory activity, than any other known archaeon; this suggests that regulatory activity in M. kandleri is highly reduced, and response pathways may be relatively few. [3]
In addition, M. kandleri codes for an unusually high proportion of negatively charged amino acids in its proteins, which contribute to the microbe’s high proportion of negatively charged residues that, as mentioned previously, is involved in conveying high salt tolerance.
The G+C content is about 60%, which is unusually high compared to most other methanogens, which average only 33% G+C . [1]
Novel proteins and features
A number of novel proteins and other compounds have been isolated from M. kandleri , and are of interest in perhaps shedding light on both its phylogenetic status and its highly thermophilic, halophilic existence. Many hyperthermophilic proteins and enzymes have been described from M. kandleri , and some have found applications in the world of biotechnology. Some of the more interesting discoveries, in terms of either evolutionary significance or possible applications, are outlined in this section.
Novel Psuedomurien Variate
Many archaeons possess a cell wall consisting of pseudomurein. Also called pseudopeptidoglycan, pseudomurein acts much as peptidoglycan does in bacterial cell walls, forming a thick, multi-layered envelope around the cell. Ordinarily, pseudomurein is made up of N-acetylglucosamine and N-acetyltalosaminuronic acid. However, Methanopyrus kandleri substitutes L-orthinine where N-acetylglucosamine ordinarily would go.
Novel Core Lipid
A number of novel lipids have been isolated from M. kandleri. The figure at right shows the structure of one of these lipids, 2,3-di- O-phytanyl-sn-glycerophosphocholine (Fig. 3). In addition, the microbe was found to utilize a novel core lipid. This lipid, an unsaturated, ether-linked lipid identified as 2,3-di-O-geranylgeranyl-sn-glycerol, is of a type of lipids (terpenoid lipids) thought to be a ‘primitive feature’ of many microbes. The presence of this terpenoid lipid may, in combination with other evidence (as detailed later in this article), provide some evidence for the claim that M. kandleri is of an early diverging branch from the Euryarchaeota group. [12]
Novel DNA-binding proteins
Sequence analysis of the M. kandleri genome shows a relative deficiency in this microbe of the helix-turn-helix and Arc-MetJ type DNA binding proteins that are usually found in abundance across both archaeons and bacteria. Instead, M. kandleri appears to utilize (at least) 3 novel DNA binding proteins. All are relatively small molecules, with molecular weights of 7, 10, and 30 kDa. The structure of the 7 kDa binding protein has been determined to be dimeric, consisting of a larger alpha-helical protein associated with a smaller, N-terminal beta-strand. Structural properties, as well as apparent regulatory similarities, suggest that this binding protein may be a relative of the ribbon-helix-helix family of transcription factors that are involved in DNA binding and gene regulation in bacteria, notably Pseudomonas bacteria. [10]
Hetero-dimeric reverse DNA gyrase
Reverse DNA gyrases, which positively supercoil DNA when activated by ATP, have been found to be common across nearly all hyperthermophiles, both archaeons and bacteria. However, the M. kandleri DNA reverse gyrase is the first hetero-dimeric DNA reverse gyrase to be described. The protein consists of two sub-units—a larger (138 kDa) subunit that associates with ATP and hydrolyses it to release energy; and a smaller (43 kDa) sub-unit that binds covalently to DNA as the protein works to create supercoils. The presence of these two sub-units within the gyrase, as opposed to having a single unit protein, is thought to help stabilize the protein at the incredibly high temperatures M. kandleri is subject to. In addition, high temperatures appear to be required for the two sub-units to form the stable heterodimer observed. [9]
Novel and modified RNA nucleosides
M. kandleri shows an unusual and highly diverse pattern of post-transcriptional modifications to tRNA nucleosides—4 entirely novel nucleosides have been isolated from this microbe. Modifications to these nucleosides effect structure, behavior, and stability of the tRNA strands each helps to form. The high degree of modification seen in M. kandleri may have to do with the extreme nature of its environment and the high demands that environment places on its proteins in terms of maintaining stability. [11]
Questions and Controversy- Placing M. kandleri within the evolutionary tree of life
Since its discovery, the placement of M. kandleri within a broader phylogenetic context has been a highly contested avenue of research. At different times, it has been thought to be many different things—a representative of a new class within Euryarchaeota , only very distantly related to its fellow methanogens [1, 2]; alternatively, quite closely related to other methanogens [3]; or, even more alternatively, quite unrelated from just about everything, and possibly a link to finding the much lauded Last Universal Common Ancestor [5]. Which one of these M. kandleri actually is, is still up to debate.
Initial Classification
The researchers who initially isolated the microbe placed it in a separate, deeper-branching, clade from other methanogens based on the following observations:
First, the G+C content of Methanopyrus was much higher than that of other known methanogens. At 60%G+C, Methanopyrus has almost twice the G+C in its genome as other methanogens. Second, the amount of homologous DNA sequences observed between Methanopyrus and other methanogens was, in their words, “insignificant.” Third, lipid analysis of the structures of Methanopyrus revealed a very different set of core lipids than what ought to be a closely related archaeons, Methanococcus . Where Methanococcus ’ core lipids are 85% comprised of cyclic dibisphytanyl tetraethers, Methanopyrus ’ core lipid was 2,3-di-O-phytanyl-glycerol, a very different type of lipid. Fourth, Methanopyrus had a higher halo-tolerance than other Methanobacteriales species. Fifth, Methanopyrus exhibited a novel type of psuedomurein in its cell wall, a variant that substituted L-ornithine for N-acetylglucosamine, and had not been observed previously. Sixth, and perhaps most convincingly, phylogenetic analysis of the microbe’s 16s RNA sequence placed it quite distantly from all known methanogens. In fact, 16s RNA analysis placed it so very distantly from other methanogens that the researchers designated it in a new class—Class Methanopyri, which they had branching off from the Phylum Euryarchaeota significantly earlier than any other known branch. [1]
Support for a separate lineage
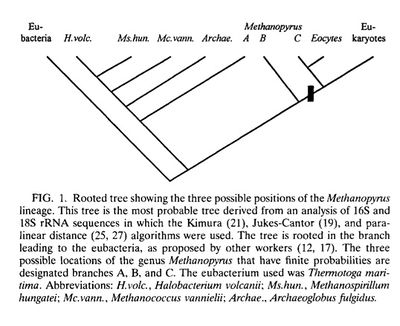
In the research since Methanopyrus ’ initial discovery in 1991, there has been some support for this theory. Analysis of M. kandleri ’s 16s RNA sequence suggests that, although a Euryarchaote, Methanopyrus shares a higher degree of relatedness to the chrenarchaeota group that seen in any other Euryarchaeote , which in turn suggests that M. kandleri may have diverged very early on. A 1996 study used a Jukes-Cantor distance projection to approach the problem qualitatively, looking at distances between 16s RNA sequences and regions of the 1-α elongation factor for M. kandleri and proposed relations, including methanogens. The tree generated by this projection again placed Methanopyrus at a deep branch within Euryarchaeota , existing either within or proximal to archaebacteria. [2] (Figure 4) A similar study using the sequences from an operon common across methanogens, coding for methyl coenzyme M reductase, also supported the conclusion that Methanopyrus stands separate from other methanogens. [6]
If this view of Methanopyrus phylogeny is correct and Methanopyrus is of a separate lineage than other methanogens, this would indicate that methanogenesis as a metabolism has arisen at least two separate times. Perhaps more importantly, because many phylogenetic analyses of Methanopyrus place it as a representative of a very deep branching group, some have looked to Methanopyrus as perhaps the most proximal known relative of the Last Universal Common Ancestor, or LUCA. Although previous efforts to locate the LUCA placed it somewhere in the bacterial realm, more recent analyses, incorporating a greater diversity of organisms, especially the previously under-represented archaeons, tell a different story. Phylogenetic distances based on tRNA sequences, which show much lower rates of horizontal transfer than protein coding genes and, as such, are more likely to generate accurate phylogenetic trees over this kind of time-frame, suggest that the LUCA falls somewhere between Euryarchaeota and Crenarchaeota —just about, in fact, where Methanopyrus kandleri seems to fit. [8] If this is the case, than M. kandleri and its genome can tell us a lot about the early divergence of life.
Support for a monophyletic view of M. kandleri and the methanogens
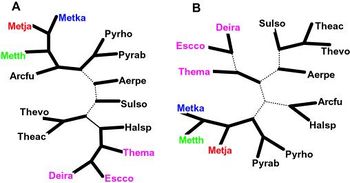
As with any radical claim in science, the proposal of a Methanopyrus-proximal LUCA met with immediate refutal. Opposing this view—opposing, in fact, the entire idea that Methanopyrus is of a different lineage than its fellow methanogens—we have a second set of research. One study used full DNA sequence data and the idea of COGs—Clusters of Orthologous Groups, sets of gene orthologs that are conserved across different phylogenetic groups—to construct a very different phylogeny, that grouped M. kandleri in one lineage with other methanogens. According to this work, about 73% of the proteins coded for in M. kandleri ’s genome are part of COGs shared across a broad spectrum of life; many of the remaining proteins are common across methanogens, which supports the proposal of a single methanogenic lineage. In the trees generated by this COG analysis, M. kandleri is shown as most proximal to either Methanococcus thermoautotrophicum or Methanococcus jannaschii.
The sequence data for these three species—Methanococcus thermoautotrophicum, Methanococcus jannaschii , and Methanopyrus kandleri —is somewhat ambiguous as to who exactly is most related to whom. All three share COGs that are common across methanogens. M. jannaschii and M. thermoautotrophicum share a partial duplication in a recently described operon. The placement and order of genes and operons is more similar between M. thermoautotrophicum and M. kandleri . Depending on what metric we pick, these three species arrange themselves into various relationships. But, despite this ongoing fine-tuning, this research makes a clear case for placing Methanopyrus within a single lineage with the methanogens. (Figure 5) [3]
Further muddying the waters, it has been suggested that many of the phylogenetic analyses that have placed M. kandleri in a separate lineage may be reflective not of an ancient divergence but of an unusually high mutation rate. Hyperthermophilic conditions such as those that M. kandleri thrives in are known to effect the stability of DNA secondary structures, causing hyperthermophilic microorganisms to exhibit higher than normal rates of sequence mutation. So one school of thought would hold that this relative genetic instability would skew any genetic distance analyses of M. kandleri towards predicting a much earlier divergence than actually occurred. On the other hand, mechanisms have been observed in many non-archaeal hyperthermophiles that mitigate wholly or entirely this effect, lowering their mutation rates to something more comparable to non-hyperthermophiles. These mechanisms, or similar mechanisms, may be in effect in M. kandleri-- however, because the DNA repair genes that encode these mechanisms are not well known in archaea, we just don't know at this point whether this is the case. (14) As seems to be often the case with M. kandleri, we are left questioning.
So—what to do with Methanopyrus, in the end? Is it a radically new lineage of methanogen, presenting new insight into the last universal origins of life itself? Is it nothing so bold, just another methanogen? With evidences that seem to point in all directions, this question is not yet resolved, and may not be any time soon. Which evidences do we choose? Do we base our phylogenetic analysis off of 16s RNA? Whole genome analysis? tRNA sequences? Whichever way we do it, we seem to get a different result. The paradox of M. kandleri remains.
References
[6] 1. Margit Kurr, Robert Huber, Helmut Konig, Holger W. Jannasch, Hans Fricke, Antonio Trineone, Jakob K. Kristjansson, and Karl O. Stetter. 1991. Methanopyrus kandleri ,gen. and sp. nov. represents a novel group of hyperthermophilic methanogens, growing at 110° C. Arch Microbiol 156:239—247.
[7] 2. Rivera, M.C., and Lake, J. 1996. The Phylogeny of Methanopyrus kandleri . IJSEM 46: 348-351.
[8] 3. Slesarev AI, Mezhevaya KV, Makarova KS, Polushin NN, Shcherbinina OV, et al. 2002. The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc Natl Acad Sci U S A 99: 4644–4649.
[9] 4. Simone Schopf, Gerhard Wanner, Reinhard Rachel, and Reinhard Wirth. 2008. An archaeal bi-species biofilm formed by Pyrococcus furiosus and Methanopyrus kandleri . Archives of Microbiology 190: 371-377.
[10] 5. Zhiliang Yu, and Shanxiang Xu. 2011. Search for a Methanopyrus -proximal last universal common ancestor based on comparative-genomic analysis. Annals of Microbiology 61: 397-401.
[11] 6. JORK NOLLING, AMY ELFNER, JOHN R. PALMER, VANESSA J. STEIGERWALD, TODD D. PIHL, JAMES A. LAKE, AND JOHN N. REEVE. 1996. Phylogeny of Methanopyms kandleri Based on Methyl Coenzyme M Reductase Operons. INTERNATIONAL JOURNAL oF SYSTEMATIC BACTERIOLOGY. 46: 1170-1173.
[12] 7. Zhiliang Yu, Ken Takai, Alexei Slesarev, Hong Xue and J. Tze-Fei Wong. 2009. Search for Primitive Methanopyrus Based on Genetic Distance Between Val- and Ile-tRNA Synthetases. Journal of Molecular Evolution 69: 386-394
[13] 8. S. Burggraf, K.O. Stetter, P. Rouviere, C.R. Woese. 1991. Methanopyrus kandleri : An Archaeal Methanogen Unrelated to all Other Known Methanogens. Systematic and Applied Microbiology 14: 346-351.
[14] 9. Regis Krah, Mary H. O’Dea and Martin Gellert. 1997. RECONSTITUTION OF AN ACTIVE EXTREMOZYME FROM ITS TWO RECOMBINANT SUBUNITS. The Journal of Biological Chemistry, 272, 13986-13990.
[15] 10. Nikolai A. Pavlov, Dmitry I. Cherny, Igor V. Nazimov, Alexei I. Slesarev, and Vinod Subramaniam. 2002. Identification, cloning and characterization of a new DNA-binding protein from the hyperthermophilic methanogen Methanopyrus kandleri . Nucl. Acids Res. (2002) 30 (3): 685-694.
[16] 11. Sauerwald, Anselm1; Sitaramaiah, Devarasetty2; McCloskey, James A.2,3; Söll, Dieter1; Crain, Pamela F.. 2005. N6-Acetyladenosine: A new modified nucleoside from Methanopyrus kandleri tRNA 579: 2807-2810.
[17] 12. HAFENBRADL, D; KELLER, M; THIERICKE, R; STETTER, KO. 1993. A NOVEL UNSATURATED ARCHAEAL ETHER CORE LIPID FROM THE HYPERTHERMOPHILE METHANOPYRUS -KANDLERI. SYSTEMATIC AND APPLIED MICROBIOLOGY 16: 165-169.
[18] 13. MASATERU NISHIHARA, HIROYUKI MORII, KOJI MATSUNO, MAMI OHGA, KARL O. STETTER, and YOSUKE KOGA. 2002. Structural analysis by reductive cleavage with LiAlH4 of an allyl ether choline-phospholipid, archaetidylcholine, from the hyperthermophilic methanoarchaeon Methanopyrus kandleri. Archaea 1, 123-131.
[19] 14. Dennis W. Grogan. 1998. Hyperthermophiles and the problem of DNA instability. Molecular Microbiology 28(6):1043–1049.
