Pithovirus sibericum: Difference between revisions
mNo edit summary |
|||
| (25 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
==Classification== | ==Classification== | ||
Viruses; dsDNA viruses, no RNA stage; unclassified dsDNA viruses. | Viruses; dsDNA viruses, no RNA stage; unclassified dsDNA viruses.[http://www.ncbi.nlm.nih.gov/genome/?term=txid1450746] | ||
===Species=== | ===Species=== | ||
| Line 14: | Line 14: | ||
==Description and Significance== | ==Description and Significance== | ||
''Pithovirus sibericum'' is a giant DNA virus of a previously unclassified family isolated from a layer of permafrost in the Kolyma Lowland region of Siberia in 2013. The sedimentary layer from which the ''Pithovirus'' was isolated dates back 30,000 years or more. ''Pithovirus'' infects amoebae; human and animal pathogenicity has been ruled out thus far. ''Pithovirus'' is notable due to its unprecedented size compared to known viruses, its unusually small genome relative to its size, and its lack of phylogenetic relationship to any known virus family. | ''Pithovirus sibericum'' is a giant DNA virus of a previously unclassified family isolated from a layer of permafrost in the Kolyma Lowland region of Siberia in 2013. The sedimentary layer from which the ''Pithovirus'' was isolated dates back 30,000 years or more. ''Pithovirus'' infects amoebae; human and animal pathogenicity has been ruled out thus far. ''Pithovirus'' is notable due to its unprecedented size compared to known viruses, its unusually small genome relative to its size, and its lack of phylogenetic relationship to any known virus family. [http://www.pnas.org.ezproxy.lib.indiana.edu/content/111/11/4274.abstract] | ||
==Genome Structure== | ==Genome Structure== | ||
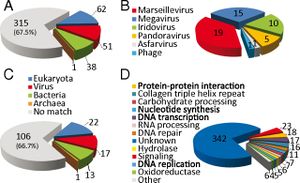
''Pithovirus sibericum’s'' genome is unexpectedly different from that of other giant DNA viruses with morphological similarity. ''Pandoravirus'', another recently discovered giant DNA virus, has the same amphora morphology and sports a large, GC-rich, 1.26 Mb genome with an estimated 2,500 protein-coding genes. By contrast, ''Pithovirus'' has an AT-rich genome with only 610,033 bp and encodes 467 proteins. Paradoxically, the giant DNA viruses with which ''Pithovirus'' shares the most genomic attributes and replication methods are morphologically dissimilar icosahedrons. Pithovirus’s genome overall structure is currently uncertain, but appears to be either linear with terminal redundancy or circular. | ''Pithovirus sibericum’s'' genome is unexpectedly different from that of other giant DNA viruses with morphological similarity. ''Pandoravirus'', another recently discovered giant DNA virus, has the same amphora morphology and sports a large, GC-rich, 1.26 Mb genome with an estimated 2,500 protein-coding genes. By contrast, ''Pithovirus'' has an AT-rich genome with only 610,033 bp and encodes 467 proteins. Paradoxically, the giant DNA viruses with which ''Pithovirus'' shares the most genomic attributes and replication methods are morphologically dissimilar icosahedrons. Pithovirus’s genome overall structure is currently uncertain, but appears to be either linear with terminal redundancy or circular.[http://www.pnas.org.ezproxy.lib.indiana.edu/content/111/11/4274.abstract 2] | ||
[[Image:Pithovirus sibericum genome.jpg|thumb|''Pithovirus sibericum'' genome composition]] | |||
The ''Pithovirus'' genome lacks genes encoding machinery associated with translation, ATP synthesis, and cell division, which confirmed that it is indeed a virus; furthermore, lack of translation machinery is consistent with cytoplasmic replication. About 21% of its genome contains a non-coding repeat that appears unique to ''Pithovirus'' as it is unrelated to repeats found in other viral genomes. Its occurrence results in a relatively low coding density compared to other viral genomes. Of the protein-coding genes, most are associated with nucleotide synthesis and DNA transcription, replication, and repair, which is a typical attribute of a large DNA virus genome. | [[File:Pithovirus phylogeny.jpg|200px|thumb|left|''Pithovirus sibericum'' phylogeny]] | ||
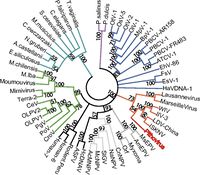
Examination of ''Pithovirus’s'' 467 protein-coding genes indicates that 67% have no homologs in any known sequence. The 33% that code for homologous proteins are equally comparable to viruses, bacteria, and eukaryotic organisms, indicating that ''Pithovirus'' has no particularly close phylogenetic relationship to any known sequenced organism. 11% of the total genome corresponds to proteins sequenced from viruses. Phylogenetic analysis of the DNA polymerase gene places ''Pithovirus'' in a clade of icosahedral large DNA virus families including ''Iridovirus'' and ''Marseillevirus'' and distinct from the morphological similar ''Pandoravirus''. Notably, this lack of close phylogenetic relationship confirms that ''Pithovirus'' belongs to a previously unclassified family of viruses. [http://www.pnas.org.ezproxy.lib.indiana.edu/content/111/11/4274.abstract 2] | |||
The ''Pithovirus'' genome lacks genes encoding machinery associated with translation, ATP synthesis, and cell division, which confirmed that it is indeed a virus; furthermore, lack of translation machinery is consistent with cytoplasmic replication. About 21% of its genome contains a non-coding repeat that appears unique to ''Pithovirus'' as it is unrelated to repeats found in other viral genomes. Its occurrence results in a relatively low coding density compared to other viral genomes. Of the protein-coding genes, most are associated with nucleotide synthesis and DNA transcription, replication, and repair, which is a typical attribute of a large DNA virus genome.[http://www.pnas.org.ezproxy.lib.indiana.edu/content/111/11/4274.abstract 2] | |||
==Cell Structure, Metabolism and Life Cycle== | ==Cell Structure, Metabolism and Life Cycle== | ||
The ''Pithovirus'' replication cycle that results in host cell lysis takes between 10-20 hours. The virus infects amoeboid cells by expelling its cork and fusing its internal lipid membrane to the host cell membrane, allowing the virus to establish virion assembly machinery within the host cell cytoplasm. The machinery establishes a "virion factory" that can be initially observed by localized clearing of cellular cytoplasmic structures. Numerous virion particles are formed simultaneously. Once virion assembly is complete, the virus is released via host cell lysis; virion particles are also found in vacuoles within the cell, indicating they | [[File:Pithovirus sibericum.jpg|200px|thumb|right|''Pithovirus sibericum'' structure]] | ||
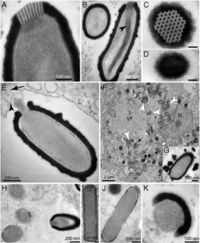
The ''Pithovirus'' virion is so far the largest virion known. It is an oblong rod, approximately 1.5 micrometers in length and 500nm in diameter, with a “cork” at its apex that is unique to this organism. The virus is encased in an external envelope and an internal lipid membrane. Within the internal membrane a tubular structure of unknown function has been observed. The cork at the apex is connected to a coil of membrane within the viral internal membrane.[http://www.pnas.org.ezproxy.lib.indiana.edu/content/111/11/4274.abstract 2] | |||
The ''Pithovirus'' replication cycle that results in host cell lysis takes between 10-20 hours. The virus infects amoeboid cells by expelling its cork and fusing its internal lipid membrane to the host cell membrane, allowing the virus to establish virion assembly machinery within the host cell cytoplasm. The machinery establishes a "virion factory" that can be initially observed by localized clearing of cellular cytoplasmic structures. Numerous virion particles are formed simultaneously. Once virion assembly is complete, the virus is released via host cell lysis; virion particles are also found in vacuoles within the cell, indicating they may also exit via exocytosis. [http://www.pnas.org.ezproxy.lib.indiana.edu/content/111/11/4274.abstract 2] | |||
==Ecology and Pathogenesis== | ==Ecology and Pathogenesis== | ||
''Pithovirus sibericum'' is a pathogen of amoebae; it was isolated by | ''Pithovirus sibericum'' is a pathogen of amoebae; it was isolated by using ''Acanthamoeba'' as bait and proved virulent after 30,000 years frozen in permafrost. The discovery of a millenia-old pathogen has important implications on the possibility of unknown viral pathogens remaining preserved in various geological structures. Effects of human industries such as mining and drilling may include the release of such unknown pathogens. Climate change in particular has the potential to alter icy geological strata and release these pathogens. Viruses play important roles in global food webs due to their effects on host populations; it has been suggested that virus-induced mortality affects whole-ecosystem functioning. The method of discovery of ''Pithovirus sibericum'' has informed the scientific community of an inexpensive way to test for the release of unknown viruses in order to mitigate possible detrimental effects on ecosystems([http://www.pnas.org.ezproxy.lib.indiana.edu/content/111/11/4274.abstract 2]); ([http://kg6ek7cq2b.search.serialssolutions.com.ezproxy.lib.indiana.edu/?&url_ver=Z39.88-2004&url_ctx_fmt=info:ofi/fmt:kev:mtx:ctx&rft_val_fmt=info:ofi/fmt:kev:mtx:journal&rft.atitle=The%20relative%20importance%20of%20viral%20lysis%20and%20nanoflagellate%20grazing%20for%20prokaryote%20mortality%20in%20temperate%20lakes&rft.aufirst=Angia%20Sriram%20Pradeep&rft.aulast=Ram&rft.date=2014&rft.eissn=1365-2427&rft.epage=311&rft.genre=article&rft.issn=0046-5070&rft.issue=2&rft.jtitle=FRESHWATER%20BIOLOGY&rft.pages=300-311&rft.spage=300&rft.stitle=FRESHWATER%20BIOL&rft.volume=59&rfr_id=info:sid/www.isinet.com:WoK:UA&rft.au=Palesse%2C%20Stephanie&rft.au=Colombet%2C%20Jonathan&rft.au=Thouvenot%2C%20Antoine&rft.au=Sime-Ngando%2C%20Telesphore&rft_id=info:doi/10.1111%2Ffwb.12265 3]) | ||
==References== | ==References== | ||
| Line 36: | Line 44: | ||
2. [http://www.pnas.org.ezproxy.lib.indiana.edu/content/111/11/4274.abstract Legendre, Matthieu; Bartoli, Julia; Shmakova, Lyubov; et al. 2014. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. PNAS, 11; 4274-4279] | 2. [http://www.pnas.org.ezproxy.lib.indiana.edu/content/111/11/4274.abstract Legendre, Matthieu; Bartoli, Julia; Shmakova, Lyubov; et al. 2014. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. PNAS, 11; 4274-4279] | ||
3. [http://kg6ek7cq2b.search.serialssolutions.com.ezproxy.lib.indiana.edu/?&url_ver=Z39.88-2004&url_ctx_fmt=info:ofi/fmt:kev:mtx:ctx&rft_val_fmt=info:ofi/fmt:kev:mtx:journal&rft.atitle=The%20relative%20importance%20of%20viral%20lysis%20and%20nanoflagellate%20grazing%20for%20prokaryote%20mortality%20in%20temperate%20lakes&rft.aufirst=Angia%20Sriram%20Pradeep&rft.aulast=Ram&rft.date=2014&rft.eissn=1365-2427&rft.epage=311&rft.genre=article&rft.issn=0046-5070&rft.issue=2&rft.jtitle=FRESHWATER%20BIOLOGY&rft.pages=300-311&rft.spage=300&rft.stitle=FRESHWATER%20BIOL&rft.volume=59&rfr_id=info:sid/www.isinet.com:WoK:UA&rft.au=Palesse%2C%20Stephanie&rft.au=Colombet%2C%20Jonathan&rft.au=Thouvenot%2C%20Antoine&rft.au=Sime-Ngando%2C%20Telesphore&rft_id=info:doi/10.1111%2Ffwb.12265 Angia Sriram Pradeep Ram, Stephanie Palesse1, Jonathan Colombet, Antoine Thouvenot and Telesphore Sime-Ngando. 2013. The relative importance of viral lysis and nanoflagellate grazing for prokaryote mortality in temperate lakes. Freshwater Biology, 59; 300-311. DOI: 10.1111/fwb.12265] | |||
==Author== | ==Author== | ||
Latest revision as of 10:59, 7 May 2014
Classification
Viruses; dsDNA viruses, no RNA stage; unclassified dsDNA viruses.[1]
Species
|
NCBI: Taxonomy |
Pithovirus Sibericum
Description and Significance
Pithovirus sibericum is a giant DNA virus of a previously unclassified family isolated from a layer of permafrost in the Kolyma Lowland region of Siberia in 2013. The sedimentary layer from which the Pithovirus was isolated dates back 30,000 years or more. Pithovirus infects amoebae; human and animal pathogenicity has been ruled out thus far. Pithovirus is notable due to its unprecedented size compared to known viruses, its unusually small genome relative to its size, and its lack of phylogenetic relationship to any known virus family. [2]
Genome Structure
Pithovirus sibericum’s genome is unexpectedly different from that of other giant DNA viruses with morphological similarity. Pandoravirus, another recently discovered giant DNA virus, has the same amphora morphology and sports a large, GC-rich, 1.26 Mb genome with an estimated 2,500 protein-coding genes. By contrast, Pithovirus has an AT-rich genome with only 610,033 bp and encodes 467 proteins. Paradoxically, the giant DNA viruses with which Pithovirus shares the most genomic attributes and replication methods are morphologically dissimilar icosahedrons. Pithovirus’s genome overall structure is currently uncertain, but appears to be either linear with terminal redundancy or circular.2
Examination of Pithovirus’s 467 protein-coding genes indicates that 67% have no homologs in any known sequence. The 33% that code for homologous proteins are equally comparable to viruses, bacteria, and eukaryotic organisms, indicating that Pithovirus has no particularly close phylogenetic relationship to any known sequenced organism. 11% of the total genome corresponds to proteins sequenced from viruses. Phylogenetic analysis of the DNA polymerase gene places Pithovirus in a clade of icosahedral large DNA virus families including Iridovirus and Marseillevirus and distinct from the morphological similar Pandoravirus. Notably, this lack of close phylogenetic relationship confirms that Pithovirus belongs to a previously unclassified family of viruses. 2
The Pithovirus genome lacks genes encoding machinery associated with translation, ATP synthesis, and cell division, which confirmed that it is indeed a virus; furthermore, lack of translation machinery is consistent with cytoplasmic replication. About 21% of its genome contains a non-coding repeat that appears unique to Pithovirus as it is unrelated to repeats found in other viral genomes. Its occurrence results in a relatively low coding density compared to other viral genomes. Of the protein-coding genes, most are associated with nucleotide synthesis and DNA transcription, replication, and repair, which is a typical attribute of a large DNA virus genome.2
Cell Structure, Metabolism and Life Cycle
The Pithovirus virion is so far the largest virion known. It is an oblong rod, approximately 1.5 micrometers in length and 500nm in diameter, with a “cork” at its apex that is unique to this organism. The virus is encased in an external envelope and an internal lipid membrane. Within the internal membrane a tubular structure of unknown function has been observed. The cork at the apex is connected to a coil of membrane within the viral internal membrane.2
The Pithovirus replication cycle that results in host cell lysis takes between 10-20 hours. The virus infects amoeboid cells by expelling its cork and fusing its internal lipid membrane to the host cell membrane, allowing the virus to establish virion assembly machinery within the host cell cytoplasm. The machinery establishes a "virion factory" that can be initially observed by localized clearing of cellular cytoplasmic structures. Numerous virion particles are formed simultaneously. Once virion assembly is complete, the virus is released via host cell lysis; virion particles are also found in vacuoles within the cell, indicating they may also exit via exocytosis. 2
Ecology and Pathogenesis
Pithovirus sibericum is a pathogen of amoebae; it was isolated by using Acanthamoeba as bait and proved virulent after 30,000 years frozen in permafrost. The discovery of a millenia-old pathogen has important implications on the possibility of unknown viral pathogens remaining preserved in various geological structures. Effects of human industries such as mining and drilling may include the release of such unknown pathogens. Climate change in particular has the potential to alter icy geological strata and release these pathogens. Viruses play important roles in global food webs due to their effects on host populations; it has been suggested that virus-induced mortality affects whole-ecosystem functioning. The method of discovery of Pithovirus sibericum has informed the scientific community of an inexpensive way to test for the release of unknown viruses in order to mitigate possible detrimental effects on ecosystems(2); (3)
References
1. National Center for Biotechnology Information txid 1450746
Author
Page authored by Jennifer Gliessman, student of Prof. Jay Lennon at IndianaUniversity.