Nitrosomonas europaea: Difference between revisions
Slonczewski (talk | contribs) |
|||
| (28 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
{{Curated}} | |||
{{Biorealm Genus}} | {{Biorealm Genus}} | ||
| Line 31: | Line 32: | ||
==Description and significance== | ==Description and significance== | ||
[[Image:Nitrosomonas-Photo_by_Stan_Watson,_Woods_Hole_Oceanographic_Institute.jpg|frame|right|Picture of Nitrosomonas europaea. Photo by Stan Watson, Woods Hole Oceanographic Institute. http://genome.jgi-psf.org/finished_microbes/niteu/niteu.home.html]] | |||
''Nitrosomonas europaea'' is a Gram-negative chemolithoautroph with the shape of bacillus. It is an ammonia-oxidizing bacterium that lives in places rich in ammonia and inorganic salt, such as soil, sewage, freshwater, the walls of buildings and on the surface of monuments. It's often found in the polluted areas where the air contains high levels of nitrogen compounds. | |||
It is important enough to have its genome sequenced because this organism plays a central role in the availability of nitrogen to plants and hence in limiting C02 fixation. These bacteria are important players in the treatment of industrial and sewage waste in the first step of oxidizing ammonia to nitrite; then other bacteria oxidize nitrite to nitrate. ''N. europaea'' is also capable of degrading a variety of halogenated organic compounds, including trichloroethylene , benzene and vinyl chloride, which may make it an attractive organism for bioremediation. | |||
==Genome structure== | |||
Its genome consists of a single circular chromosome of 2,812,094 bp. The GC skew analysis indicates that the genome is divided into two unequal replichores. Genes are distributed evenly around the genome, with approximately 47% transcribed from one strand and approximately 53% transcribed from the complementary strand. A total of 2,460 protein-encoding genes emerged from the modeling effort, averaging 1,011 bp in length, with intergenic regions averaging 117 bp. Genes necessary for the catabolism of ammonia, energy and reductant generation, biosynthesis, and CO(2) and NH(3) assimilation were identified. In contrast, genes for catabolism of organic compounds are limited. Genes encoding transporters for inorganic ions were plentiful, whereas genes encoding transporters for organic molecules were scant. Complex repetitive elements constitute ca. 5% of the genome. Among these are 85 predicted insertion sequence elements in eight different families. The strategy of ''N. europaea'' to accumulate Fe from the environment involves several classes of Fe receptors with more than 20 genes devoted to these receptors. However, genes for the synthesis of only one siderophore, citrate, were identified in the genome. This genome has provided new insights into the growth and metabolism of ammonia-oxidizing bacteria. | |||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
''Nitrosomonas europaea'' is an autotroph. It can obtain the carbon that it needs to grow by getting it from the atmosphere in a process known as "carbon fixation". Carbon fixation is the process of converting carbon in a gaseous form into carbon bound up in organic molecules. This bacterium contains "carboxysomes" (dark spots which can be seen scattered throughout the cell), which store the enzymes used to fix carbon dioxide for cell carbon. | |||
==Ecology== | ==Ecology== | ||
This nitrifying bacterium is the most studied of the ammonia-oxidizing bacteria that are participants in the biogeochemical N cycle. Nitrifying bacteria play a central role in the availability of nitrogen to plants and hence in limiting CO2 fixation. The reaction catalyzed by these bacteria is the first step in the oxidation of ammonia to nitrate. ''N.europaea'' also is capable of degrading a variety of halogenated organic compounds, including trichloroethylene, benzene, and vinyl chloride. The ability of nitrifying organisms to degrade some pollutants may make these organisms attractive for controlled bioremediation in nitrifying soils and waters. | |||
==Application to Biotechnology== | ==Application to Biotechnology== | ||
Conversion of ammonia to dinitrogen in wastewater was tried by ''Nitrosomonas europaea'' because ''Nitrosomonas europaea'' contains ammonia-oxidizing enzyme, nitrite reductase, and nitrous oxide reductase. | |||
==Current Research== | ==Current Research== | ||
The dynamics of growth and death of immobilized ''Nitrosomonas | |||
europaea'' were studied. For this, the death rate of suspended | |||
cells was determined in the absence of ammonium or oxygen by following the loss of respiration activity and by fluorescein-diacetate (FDA)/ lissamine-green staining techniques. The death rates obtained in the absence of oxygen or ammonium were incorporated in a dynamic growth model and the effects on the performance of the immobilized-cell process was illustrated by model simulations. These model simulations and experimental validation show that if decay of biomass occurs, the biomass concentration in the center of the bead decreases. As a result, the systems react slower to changes in substrate concentrations than if all cells remain viable. | |||
==References== | ==References== | ||
# Patrick Chain et al. (May 2003). "Complete Genome Sequence of the Ammonia-Oxidizing Bacterium and Obligate Chemolithoautotroph Nitrosomonas europaea". Journal of Bacteriology 185: 2759-2773. | |||
# John G. Holt, Noel R. Krieg, editor, editor-in-chief (1984) Bergey's manual of systematic bacteriology volume2 page1809- 1825 | |||
# DOE JOINT GENOME INSTITUTE - US Department of Energy Office of Science http://genome.jgi-psf.org/finished_microbes/niteu/niteu.home.html | |||
# DJW, Microbial of the Week 1999, Nirosomonas europae. http://web.umr.edu/~microbio/BIO221_1999/N_europaea.html | |||
# Nagatsuta, Midori-ku, Yokohama "Conversion of ammonia to dinitrogen in wastewater by Nitrosomonas europaea". Department of Bioengineering, Tokyo Institute of Technology http://cat.inist.fr/?aModele=afficheN&cpsidt=1056630 | |||
Wikipedia http://en.wikipedia.org/wiki/Nitrosomonas_europaea | # Wikipedia http://en.wikipedia.org/wiki/Nitrosomonas_europaea | ||
# Stacy Heather "Nitrosomonas europaea" http://web.umr.edu/~microbio/BIO221_1999/N_europaea.html | |||
Edited by student of [mailto:ralarsen@ucsd.edu Rachel Larsen] and Kit Pogliano | Edited by student of [mailto:ralarsen@ucsd.edu Rachel Larsen] and Kit Pogliano | ||
KMG | |||
Latest revision as of 21:25, 14 December 2015
A Microbial Biorealm page on the genus Nitrosomonas europaea
Classification
Higher order taxa
Domain; Phylum; Class; Order; family [Others may be used. Use NCBI link to find]
Domain: Prokaryota
Phylum: Proteobacteria
Class: Beta Proteobacteria
Order: Nitrosomonadales
Family: Nitrosomonadaceae
Genus: Nitrosomonas
Species: N. europea
Species
|
NCBI: Taxonomy |
Nitrosomonas europaea
Description and significance
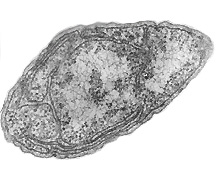
Nitrosomonas europaea is a Gram-negative chemolithoautroph with the shape of bacillus. It is an ammonia-oxidizing bacterium that lives in places rich in ammonia and inorganic salt, such as soil, sewage, freshwater, the walls of buildings and on the surface of monuments. It's often found in the polluted areas where the air contains high levels of nitrogen compounds.
It is important enough to have its genome sequenced because this organism plays a central role in the availability of nitrogen to plants and hence in limiting C02 fixation. These bacteria are important players in the treatment of industrial and sewage waste in the first step of oxidizing ammonia to nitrite; then other bacteria oxidize nitrite to nitrate. N. europaea is also capable of degrading a variety of halogenated organic compounds, including trichloroethylene , benzene and vinyl chloride, which may make it an attractive organism for bioremediation.
Genome structure
Its genome consists of a single circular chromosome of 2,812,094 bp. The GC skew analysis indicates that the genome is divided into two unequal replichores. Genes are distributed evenly around the genome, with approximately 47% transcribed from one strand and approximately 53% transcribed from the complementary strand. A total of 2,460 protein-encoding genes emerged from the modeling effort, averaging 1,011 bp in length, with intergenic regions averaging 117 bp. Genes necessary for the catabolism of ammonia, energy and reductant generation, biosynthesis, and CO(2) and NH(3) assimilation were identified. In contrast, genes for catabolism of organic compounds are limited. Genes encoding transporters for inorganic ions were plentiful, whereas genes encoding transporters for organic molecules were scant. Complex repetitive elements constitute ca. 5% of the genome. Among these are 85 predicted insertion sequence elements in eight different families. The strategy of N. europaea to accumulate Fe from the environment involves several classes of Fe receptors with more than 20 genes devoted to these receptors. However, genes for the synthesis of only one siderophore, citrate, were identified in the genome. This genome has provided new insights into the growth and metabolism of ammonia-oxidizing bacteria.
Cell structure and metabolism
Nitrosomonas europaea is an autotroph. It can obtain the carbon that it needs to grow by getting it from the atmosphere in a process known as "carbon fixation". Carbon fixation is the process of converting carbon in a gaseous form into carbon bound up in organic molecules. This bacterium contains "carboxysomes" (dark spots which can be seen scattered throughout the cell), which store the enzymes used to fix carbon dioxide for cell carbon.
Ecology
This nitrifying bacterium is the most studied of the ammonia-oxidizing bacteria that are participants in the biogeochemical N cycle. Nitrifying bacteria play a central role in the availability of nitrogen to plants and hence in limiting CO2 fixation. The reaction catalyzed by these bacteria is the first step in the oxidation of ammonia to nitrate. N.europaea also is capable of degrading a variety of halogenated organic compounds, including trichloroethylene, benzene, and vinyl chloride. The ability of nitrifying organisms to degrade some pollutants may make these organisms attractive for controlled bioremediation in nitrifying soils and waters.
Application to Biotechnology
Conversion of ammonia to dinitrogen in wastewater was tried by Nitrosomonas europaea because Nitrosomonas europaea contains ammonia-oxidizing enzyme, nitrite reductase, and nitrous oxide reductase.
Current Research
The dynamics of growth and death of immobilized Nitrosomonas
europaea were studied. For this, the death rate of suspended
cells was determined in the absence of ammonium or oxygen by following the loss of respiration activity and by fluorescein-diacetate (FDA)/ lissamine-green staining techniques. The death rates obtained in the absence of oxygen or ammonium were incorporated in a dynamic growth model and the effects on the performance of the immobilized-cell process was illustrated by model simulations. These model simulations and experimental validation show that if decay of biomass occurs, the biomass concentration in the center of the bead decreases. As a result, the systems react slower to changes in substrate concentrations than if all cells remain viable.
References
- Patrick Chain et al. (May 2003). "Complete Genome Sequence of the Ammonia-Oxidizing Bacterium and Obligate Chemolithoautotroph Nitrosomonas europaea". Journal of Bacteriology 185: 2759-2773.
- John G. Holt, Noel R. Krieg, editor, editor-in-chief (1984) Bergey's manual of systematic bacteriology volume2 page1809- 1825
- DOE JOINT GENOME INSTITUTE - US Department of Energy Office of Science http://genome.jgi-psf.org/finished_microbes/niteu/niteu.home.html
- DJW, Microbial of the Week 1999, Nirosomonas europae. http://web.umr.edu/~microbio/BIO221_1999/N_europaea.html
- Nagatsuta, Midori-ku, Yokohama "Conversion of ammonia to dinitrogen in wastewater by Nitrosomonas europaea". Department of Bioengineering, Tokyo Institute of Technology http://cat.inist.fr/?aModele=afficheN&cpsidt=1056630
- Stacy Heather "Nitrosomonas europaea" http://web.umr.edu/~microbio/BIO221_1999/N_europaea.html
Edited by student of Rachel Larsen and Kit Pogliano
KMG
