Bacteroide composition in the gut: Difference between revisions
| (179 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | {{Uncurated}} | ||
[https://en.wikipedia.org/wiki/Bacteria Bacteria] have played a significant role in [https://en.wikipedia.org/wiki/Homo_sapiens <i>Homo sapiens</i>] evolution. Only ten percent of the cells existing in the body are human. Genetically, we are about 1% human, and 99% bacterial [[#References|[10]]. | |||
[[Image:1369260_bild1_b--ckhed.jpg|thumb|300px|right|<br><b>Figure 1. </b><i>Bacteroides thetaiotaomicron</i> in relation to food particles in the mouse intestine, provided by [http://gut.bmj.com/content/early/2011/11/23/gutjnl-2011-301104.full Larsson et al., 2012] [[#References|[7]]]] | |||
The [https://en.wikipedia.org/wiki/Large_intestine human colon] contains the majority of microorganisms in the body, and 25% of these are species of [https://en.wikipedia.org/wiki/Bacteroides Bacteroides]. Bacteroides species are anaerobic, non-spore forming, gram-negative rods that have adapted to, and now thrive in, the human gut [[#References|[8]]]. The relationship between Bacteroides and the host has recently been considered mutual, given that the relationship increases the fitness of both species. Given the long history of coevolution between microbiota and the intestine, it has now been shown that bacteroides function like a multifunctional organ that provides metabolic components we do not contain within our own [https://en.wikipedia.org/wiki/Genome genome] [[#References|[14]]]. Some of these metabolic traits include [https://en.wikipedia.org/wiki/Carbohydrate carbohydrate] fermentation. Digesting complex polysaccharides frees short peptides to be reabsorbed by the large intestine and to be used for energy by the host. Through various metabolic mechanisms, Bacteroides provide amino acids and vitamins, while simultaneously utilizing a wide range of dietary [https://en.wikipedia.org/wiki/Polysaccharide polysaccharides] for growth [[#References|[6]]]. | |||
Due to the metabolic role of Bacteroides in carbohydrate fermentation, there has been some evidence showing a relationship between diet, gut flora, and obesity. However, causation of these variables is still up for debate. Given the significant role Bacteroides play in the gut, the effect of diet on their mechanisms of action and composition overall is a relevant topic of analysis [[#References|[10]]]. How body flora composition might affect adiposity is particularly crucial in light of the drastic shift in food source in the United States and its consequent growing [http://en.wikipedia.org/wiki/Epidemiology_of_obesity obesity epidemic]. | |||
<br>< | ==Characteristics of the Gut Microbiota== | ||
Advances in proteomics and genomics have allowed us to understanding bacterial functioning in ground-breaking ways. We can now compare genetic sequencing to information from microbial database to identify homologous species. We can also look at the genetic information in species from different ecosystems to com[are and contrast their functioning. Both of these methods have been used in studying Bacteroides, and are described in depth below. This recent research has shown the significant adaptive ability of gut microbiota to thrive in the specific niches of the intestine, further supporting the evidence that our relationship with microbes has evolved over thousands of years. | |||
[[Image:Journal.pbio.0050156.g003.png|thumb|280px|right|<br><b>Figure 2. </b> Lateral Gene Transfer in Bacteroidetes Promotes Niche Specialization(A) Proportions of genes found in each genome. Non-laterally transferred genes (blue) are proportionally more often involved in metabolism. Genes assigned “Primary metabolism” (yellow) and “Protein biosynthesis” (red), are compared. Upward-pointing arrowhead indicate gene enrichment and downward-pointing arrowhead show gene depletion . Asterisks indicate statistical significance, single asterisk (*) indicates p < 0.05; double asterisks (**) indicate p < 0.01; and triple asterisks (***) indicate p < 0.001. For B,C, and D see original publication by [http://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.0050156#pbio-0050156-st001 Xu et al., 2007][[#References|[13]]]]. | |||
===Characterizing the Proteome and the Genome=== | |||
Recently, genomic and [http://en.wikipedia.org/wiki/Proteomics proteomic] analyses have provided insight into the mechanisms in which microbiota adapt and thrive within varying microenvironments. Sequencing [https://microbewiki.kenyon.edu/index.php/Bacteroides_thetaiotaomicron <i>B. thetaiotaomicron</i>] and [http://en.wikipedia.org/wiki/Bacteroides_fragilis <i>B. fragilis</i>] show notable genomic commonalities with eukayotes that allow these species to adapt. These Bacteroides, like eukaryotes, have an array of possibilities for gene expression [[#References|[13]]] contributing to their multipurpose functionality. This is in part due to that fact that a large portion of their genetic make-up consists of proteins, which, allow for selection of a variety of adaptive characteristics. | |||
===Adaptive Characteristics=== | |||
The adaptive characteristics to specific niches within the gut are extremeley variable and have been shown through comparisons to other known homologous proteins and by camparing Bacteroides to other non-gut microbiota. The specific traits identified by both these means are discussed below. | |||
In order to characterize the various functions of the Bacteroide genome, putative relevant proteins have been determined by homology to other known proteins. Of the 4779 proteins identified in the <i>B. thetaiotaomicron</i> proteome, 58% were ascribed a supposed function based on other known proteins, 18% were homologous to proteins without a known function, and 24% were not homologous to any protein identified in the public domain [[#References|[13]]]. The variety of functions is presumed to aid Bacteroides species in multiple ways: the proteome induces the mechanism by which some Bacteroides acquire and hydrolyze otherwise indigestible dietary polysaccharides, and includes an environment-sensing apparatus with extracytoplasmic function [http://en.wikipedia.org/wiki/Sigma_factor sigma factors] and one- and two-component signal transduction systems [[#References|[13]]] | |||
<i>B. thetaiotaomicron</i> contains the capacity to utilize [http://en.wikipedia.org/wiki/Glycan glycans] derived from the colon. The majority (61%) of the necessary enzymes (glycohyrolases) potentially exist in the periplasm, and are necessary to the polysaccharide fermentation process that increases energy availability to the host by freeing fatty acids to be reabsorbed by the intestine. Other extracellular proteins have been shown to facilitate the binding of starches to the cell surface to be digested by other extracellular enzymes [[#References|[13]]]. | |||
< | Additionally, <i>B. thetaiotaomicron</i> was shown to contain a significant population of ECF-sigma factors, which according to other identified homologous genes, are cotranscribed with regulatory proteins that respond to external environmental stimulus and act on gene expression accordingly [[#References|[13]]]. | ||
The majority of one-component signal transduction systems were shown to be similar to other genes responsible for nutrient utilization, providing further evidence of Bacteroides’ genetically evolved fitness advantage in nutrient acquisition[[#References|[13]]]. | |||
<b>Bacteroide characteristics found in comparisons with non-gut bacteria:</b> | |||
< | |||
== | Other studies have confirmed and contributed to the understanding of Bacteroide adaptive traits and functionality through comparisons with non-gut microbiota. Xu and associates [[#References|[15]]] sequenced the genomes of two gut-dwelling Bacteroide species <i>Bacteroides vulgatus</i> and <i>Bacteroides distasonis</i> and compared their common orthologs to non-gut Bacteroides <i>Porphyromonas gingivalis</i> W83 (found in human mouth) and <i>Cytophaga hutchinsonii</i> ATCC 33406 (found in soil) (Figure 2). | ||
<br> | |||
It was shown that factors such as [http://en.wikipedia.org/wiki/Horizontal_gene_transfer lateral gene transfer], mobile elements, and [http://en.wikipedia.org/wiki/Gene_amplification gene amplification] allow Bacteroides to vary the surface of their cells, sense the environment, and gain access to nutrients in the intestine (Figure 2). The authors concluded that these processes have allowed the symbiotic relationship between Bacteroidetes and the human gut environment to thrive. | |||
<b>Summary:</b> | |||
To conclude, through genomic and proteomic sequencing, some of the mechanism underlying the adaptive capabilities of the Bacteroide species in the human gut have been identified. The expansive array of proteins available to them mediates their ability to respond to their environment, thrive within the rapidly changing microenvironment of the colon, and acquire and ferment complex carbohydrates for their own energy source as well as for the host [[#References|[13]]]. In studying how various factors (i.e. diet, stress, or antibiotic consumption) affect the microbiome, the adaptational activity of Bacteroidetes can contribute to the understanding of what cellular processes are specifically affected by external factors affecting the composition of gut flora [[#References|[15]]]. | |||
==Changes in Microbiota Compostion Associated with Diet== | |||
Understanding the taxonomy and identification of the microbiota in the gut has increased substantially in recent years. The study of [http://en.wikipedia.org/wiki/Enterotype enterotypes] is at the forefront of this research. We each have specific compositions of bacteria living with us, and it seems what we eat and where we are from hold significant roles in determining the species that live within us! Additionally, these findings can help us understand why certain populations obtain ailments that others do not. The answer could be in population-specific enterotypes. | |||
===An Overview of Enterotypes=== | |||
<br>In 2011, Arumugam and colleagues identified three clusters of microbiota species, termed enterotypes, defined as “densely populated areas in a multi-dimensional space of community composition” (p. 177) [[#References|[1]]]. Type 1 is characterized by high levels of Bacteroides, type 2 is dominated by [http://en.wikipedia.org/wiki/Prevotella Prevotella], and [http://en.wikipedia.org/wiki/Ruminococcus Ruminococcus] is prevalent in type 3. In accordance with other genomic and proteomic analysis, the Bacteroides and Parabacteroides found in the type 1 enterotype fermented carbohydrates and proteins by the use of galactosidases, hexosaminidases, and proteases enzymes for degradation as well as through [http://en.wikipedia.org/wiki/Glycolysis glycolysis] and [http://en.wikipedia.org/wiki/Pentose_phosphate_pathway pentose phosphate pathways]. It was shown that enterotype composition is species driven, however, identification of molecular functioning of each species is necessary to determine health differences between hosts with varying enterotypes. Age, body mass index, gender, or nationality showed no clear causation to enterotype composition. | |||
[[File: F2.medium.gif|thumb|400px|left|Figure 3. (A) Bacterial taxon for 15 most abundant types in human stool data and pooled sewage data. (B) Box plot showing six most abundant bacterial families in human stool data both from human stool data and sewage stool data sets when non-human oligotypes were removed. Circles represent mean values, and vertical lines show first and third quartiles. Source: [http://mbio.asm.org/content/6/2/e02574-14.full Newton et al., 2015] [[#References|[3]]]]. | |||
Another study founded categorical bacterial families among different U.S. populations. The study found sewage samples to reflect the bacterial composition of human stool samples in the United States (Figure 3). While the representation of <i>Lachnospiraceae, Prevotellaceae, and Bacteroidaceae</i> seemed to differ between sewage and human stool data, the oligotype relative abundance profiles were similar. Furthermore, the results reflected thos of Arumugam and associates, showing similar U.S. specific bacterial composition characterized by enterotype 1[[#References|[3]]]]. | |||
To reiterate, several studies have confirmed differences in types of bacteria in the gut from people of various geographic locations. | |||
===Dietary Changes and Enterotype Composition=== | |||
Whether or not diet is the ultimate causative factor in determining enterotype differences is not yet clear. However, some studies show long-term dietary changes alter the microbiota of the gut. A study by Wu and colleagues (2011) analyzed fecal samples, using diet inventories and 16S rDNA sequencing, from 98 individuals in order to determine the effect of diet on enterotype identity and microbiome composition [[#References|[14]]]. The authors concluded that while microbiome composition changed within 24 hours, enterotype identity shifts are associated with long-term dietary changes. Composition of enterotypes, particularly protein and animal fat (Bacteroides) versus carbohydrates (Prevotella), were most affected in long-term analysis. Their findings provide evidence that diet affects the amount of bacteroides living in the gut, and that the change is time-dependent. | |||
Another study also showed a possible correlation between long-term diet and enterotype composition[[#References|[4]]]. De Filippo et al., in 2010, used high-throughput [http://en.wikipedia.org/wiki/16S_ribosomal_RNA 16S rDNA] sequencing and biochemical analyses to assess microbiota differences between children eating a fiber-rich, high-carbohydrate diet in [http://en.wikipedia.org/wiki/Burkina_Faso Burkina Faso] and European children fed diets high in fat and protein. The European children showed high levels of Bacteroides, while the Burkina Faso children contained microbiomes enriched by the Prevotella enterotype, known to contain genes that allow for [http://en.wikipedia.org/wiki/Xylan xylan] and [http://en.wikipedia.org/wiki/Cellulose cellulose] hydrolysis. This genetic advantage may be completely lacking in children fed western diets. While there are many distinguishing factors between the two populations that might account for enterotype differences, diet is an obvious possible causative factor given what is known about the metabolic properties of bacterial species in the gut. | |||
===Possible Associations Between Enterotype and Disease=== | |||
The authors of the Africa/Europe comparison hypothesize that diseases associated with enterotype composition may be treated by long-term dietary changes. It is also suggested that preservation of human communities in isolated areas is important, in that they may contain fitness advantages due to ancient coevolutionary symbioses between themselves and their microbiomes. These fitness advantages may explain why these communities are resistant to certain diseases [[#References|[4]]]. <br> | |||
==Bacteroides and Obesity== | |||
It is unclear whether or not the microbiota of the gut have any causative relationship with obesity. Some studies suggests that carbohydrate or protein based diets impact both the microbiota and adiposity[[#References|[7]]][[#References|[10]]]. While others show not evidence of bacterial changes in relation to body weight[[#References|[1]]][[#References|[4]]]. There is some research that suggests certain bacterial modulation of the inflammation process is a process important to weight maintanence[[#References|[2]]]. The details of these studies are described below. | |||
===Microbiota May Influence BMI=== | |||
<br>Several studies have shown relationships between diet, obesity, and microbiome composition. One study analyzed, via genetic sequencing, the bacterial contents of fecal samples from 12 “obese” and 5 “lean” (according to the [http://en.wikipedia.org/wiki/Body_mass_index BMI] scale) individuals [[#References|[8]]]. Obese individuals were found to have 20% more Firmicutes species and almost 90% less Bacteroidetes compared to the lean group. Furthermore, after one year spent on a low-carbohydrate or low-fat diet, obese individuals showed a significant change in microbiota composition after shedding 25% of their body weight. Proportions of Bacteriodetes increased while the Firmicutes population decreased after the year-long diet and weight loss. The results by Ley and associates provide evidence toward an inverse relationship between adiposity and Bacteroides proportions in the gut. Their evidence also suggests dietary therapeutic techniques toward treating or preventing obesity-associated disease. However, the small sample size of the study lens itself to critical debate in its applicability to the larger population. | |||
[[ Image:s12866-014-0311-6-2.jpg|thumb|280px|right|<br>Figure 4. Principal correspondence analysis of UniFrac distances. Difference in gut microbiota according to (A) geographic location, (B) BMI, (C) Gender. Source: [http://www.biomedcentral.com/1471-2180/14/311 Ecobar et al., 2014][[#References|[6]]]] | |||
A study by Turnbaugh (2009) [[#References|[10]]] showed parallel results to those of Ley, Turnbaugh, and Gordon (2006)[[#References|[8]]]The authors successfully mimicked human gut environments in mice by transplanting fecal matter into germ-free C57BL/6J (“humanized”) mice by metagenomic analysis of their bacterial colonization. Transitioning from a low-fat, plant-rich diet to a high-sugar “western” diet significantly changed the contents of the host’s microbiota in a single day. These changes altered metabolic pathways and gene expression of the microbiome, and eventually increased adiposity. Adiposity fluctuated depending on microbiota transplantation. Humanized mice fed the western diet showed higher levels of Firmicute species and Erysipelotrichi and Bacilli species and less representation of Bacteroides. The pathways identified in the microbiome of the lean mice included glycan degradation and fatty acid metabolism, the same pathways previously assigned to Bacteroides. These findings support the hypothesis that Bacteroides may contribute to lower body mass, and that microbiota composition in the colon is responsive to changes in diet. | |||
===Possible Causation=== | |||
Some empirical evidence suggests high-fat diets and their effects on microbiota composition negatively impact gut permeability, leading to intestinal inflammation through translocation of [http://en.wikipedia.org/wiki/Toll-like_receptor Tlr ligands]. Cani’s data suggests increased [https://en.wikipedia.org/wiki/Lipopolysaccharide_binding_protein plasma LPS] induces [http://en.wikipedia.org/wiki/Inflammation inflammation], which was shown to contribute to weight gain. During the inflammation process, the proportion of Bacteroides increased, providing one explanatory factor for diet-induced shifts in microbiota composition [[#References|[2]]]. | |||
===BMI and Bacteroid Composition May be Unrelated=== | |||
A correlation between decreased proportions of Bacteroides in the microbiota and obesity has not been found in every investigation on the topic. Arumugam and colleagues showed no correlation between type 1 enterotype and leanness [[#References|[1]]]. SImilarly, Duncan and colleagues found no evidence of differences in Bacteroide proportions in fecal samples between obese and non-obese subjects, and weight loss dieting did not alter their original observations [[#References|[4]]]. | |||
==Geography Influences Microbiota Composition== | |||
In a study by Escobar and colleagues, geography play an important role in determining microbiota composition. Through rDNA pyrosequencing of the gut microbiota of Columbian adults and comparing it with results from Americans, Europeans, and Japanese and South Koreans, they found that geography indicated more variance in microbiota composition than BMI or gender (Figure 4). They also found that Firmicutes presence was lessened with increasing BMI, however, they found no change in levels of Bacteroidetes. The same result was found in Americans, while the Japanese showed a higher proportion of Actinobacteria. | |||
These results show that factors contributing to proportions of Bacteroide composition in the gut are complex. Indeed, geography, diet, and body mass have each been shown independently to have an effect, however, many of the results negate previous findings. More research should be conducted to accumulate evidence for some of these hypotheses. | |||
==Further Reading== | ==Further Reading== | ||
[ | Study of functions of Bacteroides has expanded dramatically in recent years. Bacteroide fragilis has been found to regulate cell surface adaptation through DNA inversions, help regulate the immune system, and modulate inflammation, to name a few. It seems that we can no longer study the gut, or the whole body for that matter, without also studying its microbial communities. They have evolved with us since our origins, therefore we cannot think of ourselves in isolation. Furthermore, our actions, birthplace, and ecosystem affect these communities. Perhaps an understanding of them will reveal how our lifestyles can benefit the commensals that benefit us. Future findings will further develop personalized medicine, a strategy to treat ill health in light of individual physiological characteristics, including bacteroide composition in the gut. might Below are some relevant articles worth exploring for more information. | ||
[http://www.pnas.org/content/101/41/14919: Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation]--Proceedings of the National Academy of Sciences | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1774411/ Microbes, immunoregulation, and the gut]--British Society of Gastroentemology | |||
[http://www.ncbi.nlm.nih.gov/pubmed/15699220: Origins and evolution of the Western diet: health implications for the 21st century]--American Society for Clinical Nutrition | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1774052/Probiotics in inflammatory bowel disease: is it all gut flora modulation?]--British Society of Gastroentemology | |||
==References== | ==References== | ||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2894525/ Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The Effect of Diet on the Human Gut Microbiome: A Metagenomic Analysis in Humanized Gnotobiotic Mice. Science translational medicine. 2009;1(6):6ra14.] | 1. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3728647/ Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174-180. doi:10.1038/nature09944.] | ||
2. [http://www.eurekaselect.com/69216/article Cani, Patrice D., and Nathalie M. Delzenne. "The Role of the Gut Microbiota in Energy Metabolism and Metabolic Disease." Current Pharmaceutical Design 15.13 (2009): 1546-558. Web.] | |||
3. [http://www.biomedcentral.com/1471-2180/14/311 JS, Klotz B, Valdes BE, Agudelo GM. The gut microbiota of Colombians differs from that of Americans, Europeans and Asians. BMC Microbiol. 2014;14(1):311] | |||
4. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2930426/ De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(33):14691-14696.] | |||
5. [http://www.ncbi.nlm.nih.gov/pubmed/18779823 Duncan S, Lobley G, Flint H, et al. Human colonic microbiota associated with diet, obesity and weight loss. International Journal Of Obesity. November 2008;32(11):1720-1724. Available from: Academic Search Premier, Ipswich, MA. Accessed March 21, 2015.] | |||
6. [http://www.ncbi.nlm.nih.gov/pubmed/12055347 Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283-307.] | |||
7. [http://wlab.gu.se/digitalAssets/1377/1377805_larsson_et_al_gut_2012.pdf Larsson, E., V. Tremaroli, Y. S. Lee, O. Koren, I. Nookaew, A. Fricker, J. Nielsen, R. E. Ley, and F. Backhed. "Analysis of Gut Microbial Regulation of Host Gene Expression along the Length of the Gut and Regulation of Gut Microbial Ecology through MyD88." Gut 61.8 (2012): 1124-131. Gut. BMJ, 3 July 2012. Web. 20 Mar. 2015.] | |||
8. [http://www.ncbi.nlm.nih.gov/pubmed/17183309?dopt=Abstract&holding=npg Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022-3.] | |||
9. [http://mbio.asm.org/content/6/2/e02574-14.full RJ, Mclellan SL, Dila DK, et al. Sewage reflects the microbiomes of human populations. MBio. 2015;6(2):e02574.] | |||
10. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2894525/ Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The Effect of Diet on the Human Gut Microbiome: A Metagenomic Analysis in Humanized Gnotobiotic Mice. Science translational medicine. 2009;1(6):6ra14.] | |||
11. [http://www.ncbi.nlm.nih.gov/pubmed/17934076 Wexler HM. Bacteroides: the Good, the Bad, and the Nitty-Gritty. Clinical Microbiology Reviews. 2007;20(4):593-621. doi:10.1128/CMR.00008-07.] | |||
[http://www.ncbi.nlm.nih.gov/ | 12. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3368382/#R13 Wu GD, Chen J, Hoffmann C, et al. Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes. Science (New York, N.y). 2011;334(6052):105-108. doi:10.1126/science.1208344.] | ||
[http://www.ncbi.nlm.nih.gov/pubmed/12663928 Xu J, Bjursell MK, Himrod J, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299(5615):2074-6.] | 13. [http://www.ncbi.nlm.nih.gov/pubmed/12663928 Xu J, Bjursell MK, Himrod J, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299(5615):2074-6.] | ||
[http://ry6af4uu9w.search.serialssolutions.com/OpenURL_local?sid=Entrez:PubMed&id=pmid:17934076 Xu J, Gordon JI. Honor thy symbionts. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(18):10452-10459.] | 14. [http://ry6af4uu9w.search.serialssolutions.com/OpenURL_local?sid=Entrez:PubMed&id=pmid:17934076 Xu J, Gordon JI. Honor thy symbionts. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(18):10452-10459.] | ||
15. [http://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.0050156#pbio-0050156-st001 Xu J, Mahowald MA, Ley RE, et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007;5(7):e156.] | |||
<!--Do not remove this line--> | <!--Do not remove this line--> | ||
Edited by Maggie Schein, a student of [http://www.jsd.claremont.edu/faculty/profile.asp?FacultyID=254 | Edited by Maggie Schein, a student of [http://www.jsd.claremont.edu/faculty/profile.asp?FacultyID=254 Suzanne Kern] in BIOL168L S2 (Microbiology) in [http://www.jsd.claremont.edu/ The Keck Science Department of the Claremont Colleges] Spring 2014. | ||
<!--Do not edit or remove this line-->[[Category:Pages edited by students of Nora Sullivan at the Claremont Colleges]] | <!--Do not edit or remove this line-->[[Category:Pages edited by students of Nora Sullivan at the Claremont Colleges]] | ||
Latest revision as of 07:55, 14 April 2015
Bacteria have played a significant role in Homo sapiens evolution. Only ten percent of the cells existing in the body are human. Genetically, we are about 1% human, and 99% bacterial [10.
Figure 1. Bacteroides thetaiotaomicron in relation to food particles in the mouse intestine, provided by Larsson et al., 2012 [7
The human colon contains the majority of microorganisms in the body, and 25% of these are species of Bacteroides. Bacteroides species are anaerobic, non-spore forming, gram-negative rods that have adapted to, and now thrive in, the human gut [8]. The relationship between Bacteroides and the host has recently been considered mutual, given that the relationship increases the fitness of both species. Given the long history of coevolution between microbiota and the intestine, it has now been shown that bacteroides function like a multifunctional organ that provides metabolic components we do not contain within our own genome [14]. Some of these metabolic traits include carbohydrate fermentation. Digesting complex polysaccharides frees short peptides to be reabsorbed by the large intestine and to be used for energy by the host. Through various metabolic mechanisms, Bacteroides provide amino acids and vitamins, while simultaneously utilizing a wide range of dietary polysaccharides for growth [6].
Due to the metabolic role of Bacteroides in carbohydrate fermentation, there has been some evidence showing a relationship between diet, gut flora, and obesity. However, causation of these variables is still up for debate. Given the significant role Bacteroides play in the gut, the effect of diet on their mechanisms of action and composition overall is a relevant topic of analysis [10]. How body flora composition might affect adiposity is particularly crucial in light of the drastic shift in food source in the United States and its consequent growing obesity epidemic.
Characteristics of the Gut Microbiota
Advances in proteomics and genomics have allowed us to understanding bacterial functioning in ground-breaking ways. We can now compare genetic sequencing to information from microbial database to identify homologous species. We can also look at the genetic information in species from different ecosystems to com[are and contrast their functioning. Both of these methods have been used in studying Bacteroides, and are described in depth below. This recent research has shown the significant adaptive ability of gut microbiota to thrive in the specific niches of the intestine, further supporting the evidence that our relationship with microbes has evolved over thousands of years.
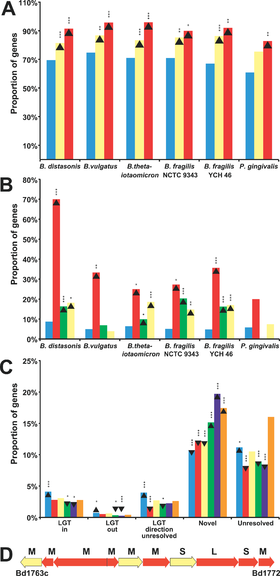
Figure 2. Lateral Gene Transfer in Bacteroidetes Promotes Niche Specialization(A) Proportions of genes found in each genome. Non-laterally transferred genes (blue) are proportionally more often involved in metabolism. Genes assigned “Primary metabolism” (yellow) and “Protein biosynthesis” (red), are compared. Upward-pointing arrowhead indicate gene enrichment and downward-pointing arrowhead show gene depletion . Asterisks indicate statistical significance, single asterisk (*) indicates p < 0.05; double asterisks (**) indicate p < 0.01; and triple asterisks (***) indicate p < 0.001. For B,C, and D see original publication by Xu et al., 2007[13
.
Characterizing the Proteome and the Genome
Recently, genomic and proteomic analyses have provided insight into the mechanisms in which microbiota adapt and thrive within varying microenvironments. Sequencing B. thetaiotaomicron and B. fragilis show notable genomic commonalities with eukayotes that allow these species to adapt. These Bacteroides, like eukaryotes, have an array of possibilities for gene expression [13] contributing to their multipurpose functionality. This is in part due to that fact that a large portion of their genetic make-up consists of proteins, which, allow for selection of a variety of adaptive characteristics.
Adaptive Characteristics
The adaptive characteristics to specific niches within the gut are extremeley variable and have been shown through comparisons to other known homologous proteins and by camparing Bacteroides to other non-gut microbiota. The specific traits identified by both these means are discussed below.
In order to characterize the various functions of the Bacteroide genome, putative relevant proteins have been determined by homology to other known proteins. Of the 4779 proteins identified in the B. thetaiotaomicron proteome, 58% were ascribed a supposed function based on other known proteins, 18% were homologous to proteins without a known function, and 24% were not homologous to any protein identified in the public domain [13]. The variety of functions is presumed to aid Bacteroides species in multiple ways: the proteome induces the mechanism by which some Bacteroides acquire and hydrolyze otherwise indigestible dietary polysaccharides, and includes an environment-sensing apparatus with extracytoplasmic function sigma factors and one- and two-component signal transduction systems [13]
B. thetaiotaomicron contains the capacity to utilize glycans derived from the colon. The majority (61%) of the necessary enzymes (glycohyrolases) potentially exist in the periplasm, and are necessary to the polysaccharide fermentation process that increases energy availability to the host by freeing fatty acids to be reabsorbed by the intestine. Other extracellular proteins have been shown to facilitate the binding of starches to the cell surface to be digested by other extracellular enzymes [13].
Additionally, B. thetaiotaomicron was shown to contain a significant population of ECF-sigma factors, which according to other identified homologous genes, are cotranscribed with regulatory proteins that respond to external environmental stimulus and act on gene expression accordingly [13].
The majority of one-component signal transduction systems were shown to be similar to other genes responsible for nutrient utilization, providing further evidence of Bacteroides’ genetically evolved fitness advantage in nutrient acquisition[13].
Bacteroide characteristics found in comparisons with non-gut bacteria:
Other studies have confirmed and contributed to the understanding of Bacteroide adaptive traits and functionality through comparisons with non-gut microbiota. Xu and associates [15] sequenced the genomes of two gut-dwelling Bacteroide species Bacteroides vulgatus and Bacteroides distasonis and compared their common orthologs to non-gut Bacteroides Porphyromonas gingivalis W83 (found in human mouth) and Cytophaga hutchinsonii ATCC 33406 (found in soil) (Figure 2).
It was shown that factors such as lateral gene transfer, mobile elements, and gene amplification allow Bacteroides to vary the surface of their cells, sense the environment, and gain access to nutrients in the intestine (Figure 2). The authors concluded that these processes have allowed the symbiotic relationship between Bacteroidetes and the human gut environment to thrive.
Summary:
To conclude, through genomic and proteomic sequencing, some of the mechanism underlying the adaptive capabilities of the Bacteroide species in the human gut have been identified. The expansive array of proteins available to them mediates their ability to respond to their environment, thrive within the rapidly changing microenvironment of the colon, and acquire and ferment complex carbohydrates for their own energy source as well as for the host [13]. In studying how various factors (i.e. diet, stress, or antibiotic consumption) affect the microbiome, the adaptational activity of Bacteroidetes can contribute to the understanding of what cellular processes are specifically affected by external factors affecting the composition of gut flora [15].
Changes in Microbiota Compostion Associated with Diet
Understanding the taxonomy and identification of the microbiota in the gut has increased substantially in recent years. The study of enterotypes is at the forefront of this research. We each have specific compositions of bacteria living with us, and it seems what we eat and where we are from hold significant roles in determining the species that live within us! Additionally, these findings can help us understand why certain populations obtain ailments that others do not. The answer could be in population-specific enterotypes.
An Overview of Enterotypes
In 2011, Arumugam and colleagues identified three clusters of microbiota species, termed enterotypes, defined as “densely populated areas in a multi-dimensional space of community composition” (p. 177) [1]. Type 1 is characterized by high levels of Bacteroides, type 2 is dominated by Prevotella, and Ruminococcus is prevalent in type 3. In accordance with other genomic and proteomic analysis, the Bacteroides and Parabacteroides found in the type 1 enterotype fermented carbohydrates and proteins by the use of galactosidases, hexosaminidases, and proteases enzymes for degradation as well as through glycolysis and pentose phosphate pathways. It was shown that enterotype composition is species driven, however, identification of molecular functioning of each species is necessary to determine health differences between hosts with varying enterotypes. Age, body mass index, gender, or nationality showed no clear causation to enterotype composition.

.
Another study founded categorical bacterial families among different U.S. populations. The study found sewage samples to reflect the bacterial composition of human stool samples in the United States (Figure 3). While the representation of Lachnospiraceae, Prevotellaceae, and Bacteroidaceae seemed to differ between sewage and human stool data, the oligotype relative abundance profiles were similar. Furthermore, the results reflected thos of Arumugam and associates, showing similar U.S. specific bacterial composition characterized by enterotype 1[3]].
To reiterate, several studies have confirmed differences in types of bacteria in the gut from people of various geographic locations.
Dietary Changes and Enterotype Composition
Whether or not diet is the ultimate causative factor in determining enterotype differences is not yet clear. However, some studies show long-term dietary changes alter the microbiota of the gut. A study by Wu and colleagues (2011) analyzed fecal samples, using diet inventories and 16S rDNA sequencing, from 98 individuals in order to determine the effect of diet on enterotype identity and microbiome composition [14]. The authors concluded that while microbiome composition changed within 24 hours, enterotype identity shifts are associated with long-term dietary changes. Composition of enterotypes, particularly protein and animal fat (Bacteroides) versus carbohydrates (Prevotella), were most affected in long-term analysis. Their findings provide evidence that diet affects the amount of bacteroides living in the gut, and that the change is time-dependent.
Another study also showed a possible correlation between long-term diet and enterotype composition[4]. De Filippo et al., in 2010, used high-throughput 16S rDNA sequencing and biochemical analyses to assess microbiota differences between children eating a fiber-rich, high-carbohydrate diet in Burkina Faso and European children fed diets high in fat and protein. The European children showed high levels of Bacteroides, while the Burkina Faso children contained microbiomes enriched by the Prevotella enterotype, known to contain genes that allow for xylan and cellulose hydrolysis. This genetic advantage may be completely lacking in children fed western diets. While there are many distinguishing factors between the two populations that might account for enterotype differences, diet is an obvious possible causative factor given what is known about the metabolic properties of bacterial species in the gut.
Possible Associations Between Enterotype and Disease
The authors of the Africa/Europe comparison hypothesize that diseases associated with enterotype composition may be treated by long-term dietary changes. It is also suggested that preservation of human communities in isolated areas is important, in that they may contain fitness advantages due to ancient coevolutionary symbioses between themselves and their microbiomes. These fitness advantages may explain why these communities are resistant to certain diseases [4].
Bacteroides and Obesity
It is unclear whether or not the microbiota of the gut have any causative relationship with obesity. Some studies suggests that carbohydrate or protein based diets impact both the microbiota and adiposity[7][10]. While others show not evidence of bacterial changes in relation to body weight[1][4]. There is some research that suggests certain bacterial modulation of the inflammation process is a process important to weight maintanence[2]. The details of these studies are described below.
Microbiota May Influence BMI
Several studies have shown relationships between diet, obesity, and microbiome composition. One study analyzed, via genetic sequencing, the bacterial contents of fecal samples from 12 “obese” and 5 “lean” (according to the BMI scale) individuals [8]. Obese individuals were found to have 20% more Firmicutes species and almost 90% less Bacteroidetes compared to the lean group. Furthermore, after one year spent on a low-carbohydrate or low-fat diet, obese individuals showed a significant change in microbiota composition after shedding 25% of their body weight. Proportions of Bacteriodetes increased while the Firmicutes population decreased after the year-long diet and weight loss. The results by Ley and associates provide evidence toward an inverse relationship between adiposity and Bacteroides proportions in the gut. Their evidence also suggests dietary therapeutic techniques toward treating or preventing obesity-associated disease. However, the small sample size of the study lens itself to critical debate in its applicability to the larger population.
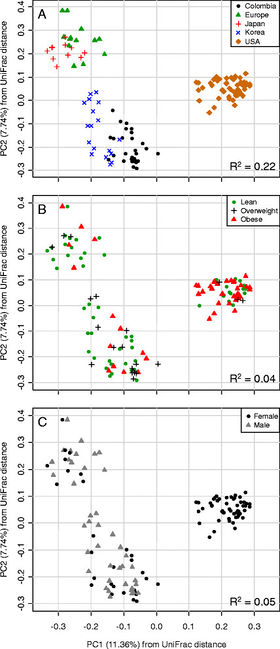
Figure 4. Principal correspondence analysis of UniFrac distances. Difference in gut microbiota according to (A) geographic location, (B) BMI, (C) Gender. Source: Ecobar et al., 2014[6
A study by Turnbaugh (2009) [10] showed parallel results to those of Ley, Turnbaugh, and Gordon (2006)[8]The authors successfully mimicked human gut environments in mice by transplanting fecal matter into germ-free C57BL/6J (“humanized”) mice by metagenomic analysis of their bacterial colonization. Transitioning from a low-fat, plant-rich diet to a high-sugar “western” diet significantly changed the contents of the host’s microbiota in a single day. These changes altered metabolic pathways and gene expression of the microbiome, and eventually increased adiposity. Adiposity fluctuated depending on microbiota transplantation. Humanized mice fed the western diet showed higher levels of Firmicute species and Erysipelotrichi and Bacilli species and less representation of Bacteroides. The pathways identified in the microbiome of the lean mice included glycan degradation and fatty acid metabolism, the same pathways previously assigned to Bacteroides. These findings support the hypothesis that Bacteroides may contribute to lower body mass, and that microbiota composition in the colon is responsive to changes in diet.
Possible Causation
Some empirical evidence suggests high-fat diets and their effects on microbiota composition negatively impact gut permeability, leading to intestinal inflammation through translocation of Tlr ligands. Cani’s data suggests increased plasma LPS induces inflammation, which was shown to contribute to weight gain. During the inflammation process, the proportion of Bacteroides increased, providing one explanatory factor for diet-induced shifts in microbiota composition [2].
A correlation between decreased proportions of Bacteroides in the microbiota and obesity has not been found in every investigation on the topic. Arumugam and colleagues showed no correlation between type 1 enterotype and leanness [1]. SImilarly, Duncan and colleagues found no evidence of differences in Bacteroide proportions in fecal samples between obese and non-obese subjects, and weight loss dieting did not alter their original observations [4].
Geography Influences Microbiota Composition
In a study by Escobar and colleagues, geography play an important role in determining microbiota composition. Through rDNA pyrosequencing of the gut microbiota of Columbian adults and comparing it with results from Americans, Europeans, and Japanese and South Koreans, they found that geography indicated more variance in microbiota composition than BMI or gender (Figure 4). They also found that Firmicutes presence was lessened with increasing BMI, however, they found no change in levels of Bacteroidetes. The same result was found in Americans, while the Japanese showed a higher proportion of Actinobacteria.
These results show that factors contributing to proportions of Bacteroide composition in the gut are complex. Indeed, geography, diet, and body mass have each been shown independently to have an effect, however, many of the results negate previous findings. More research should be conducted to accumulate evidence for some of these hypotheses.
Further Reading
Study of functions of Bacteroides has expanded dramatically in recent years. Bacteroide fragilis has been found to regulate cell surface adaptation through DNA inversions, help regulate the immune system, and modulate inflammation, to name a few. It seems that we can no longer study the gut, or the whole body for that matter, without also studying its microbial communities. They have evolved with us since our origins, therefore we cannot think of ourselves in isolation. Furthermore, our actions, birthplace, and ecosystem affect these communities. Perhaps an understanding of them will reveal how our lifestyles can benefit the commensals that benefit us. Future findings will further develop personalized medicine, a strategy to treat ill health in light of individual physiological characteristics, including bacteroide composition in the gut. might Below are some relevant articles worth exploring for more information.
Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation--Proceedings of the National Academy of Sciences
Microbes, immunoregulation, and the gut--British Society of Gastroentemology
Origins and evolution of the Western diet: health implications for the 21st century--American Society for Clinical Nutrition
in inflammatory bowel disease: is it all gut flora modulation?--British Society of Gastroentemology
References
Edited by Maggie Schein, a student of Suzanne Kern in BIOL168L S2 (Microbiology) in The Keck Science Department of the Claremont Colleges Spring 2014.
