Teixobactin: Difference between revisions
No edit summary |
No edit summary |
||
| (31 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | {{Uncurated}} | ||
The development of new antibiotics has been a difficult task further complicated by the the rapid evolution of resistant bacteria. Teixobactin is a newly discovered antibiotic that is effective against [http://en.wikipedia.org/wiki/Gram-positive_bacteria gram positive bacteria] including [http://en.wikipedia.org/wiki/Antimicrobial_resistance antibiotic-resistant] strains | [[File:Teixobactin molecule.png|thumb|300px|right|Stucture of teixobactin. By Kopiersperre via Wikimedia Commons[http://commons.wikimedia.org/wiki/File%3ATeixobactin.svg]]] | ||
The development of new antibiotics has been a difficult task further complicated by the the rapid evolution of resistant bacteria. Teixobactin is a newly discovered antibiotic that is effective against [http://en.wikipedia.org/wiki/Gram-positive_bacteria gram positive bacteria] including [http://en.wikipedia.org/wiki/Antimicrobial_resistance antibiotic-resistant] strains without evidence of resistance development.[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>] The antibiotic, isolated from the soil using a device called the iChip, works by inhibiting cell wall synthesis.[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>] | |||
==Introduction== | ==Introduction== | ||
[[Image:Teixobactin-The-Newly-Discovered-Antibiotic.png|thumb|600px|right|An infographic on teixobactin's discovery, mechanism of action, and limitations. By Andy Brunning of Compound Interest[http://www.compoundchem.com/wp-content/uploads/2015/01/Teixobactin-The-Newly-Discovered-Antibiotic.png] | [[Image:Teixobactin-The-Newly-Discovered-Antibiotic.png|thumb|600px|right|An infographic on teixobactin's discovery, mechanism of action, and limitations. By Andy Brunning of Compound Interest[http://www.compoundchem.com/wp-content/uploads/2015/01/Teixobactin-The-Newly-Discovered-Antibiotic.png]]] | ||
] | |||
< | Microorganisms have been our primary source of new antibiotics since the drugs' introduction in the 1940s.[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>] However, only about 1% of all bacterial species are readily culturable in laboratory conditions.[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>] Despite the need for new drugs, finding new antibiotics is expensive and difficult, and the identification of new, effective compounds have dramatically decreased since the mid-twentieth century.[http://www.nature.com/nature/journal/v517/n7535/full/nature14193.html <sup>2</sup>] NovoBiotic Pharmaceuticals in collaboration with the University of Bonn and Northeastern University reported in [http://en.wikipedia.org/wiki/Nature_(journal) <i>Nature</i>] the discovery of a new antibiotic, teixobactin, with properties that minimize the development of bacterial resistance.[http://static.squarespace.com/static/54878590e4b0b8980d215324/t/54ad7aaae4b09209a6148464/1420655274323/2015+january+7th+Novobiotic+press+release.pdf <sup>3</sup>] Ling <i>et al.</i> found teixobactin to be effective against gram positive bacteria, including drug-resistant strains such as methicilin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE).[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>] This antibiotic, isolated using a device called the iChip, works through inhibiting cell wall synthesis by binding to two lipid cell wall precursors, lipid II and lipid III.[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>] Teixobactin was highly effective in treating MRSA and <i>Streptococcus pneumoniae</i> in mice via injection.[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>] | ||
<br> <br> | <br> <br> | ||
==Identification== | ==Identification== | ||
Teixobactin was isolated from a new species, <i>Eleftheria terrae</i>, from soil using a [http://en.wikipedia.org/wiki/High-throughput_screening high-throughput] device called the iChip.[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>]The iChip is used to isolate previously unculturable bacteria by growing microbes [http://en.wikipedia.org/wiki/In_situ in situ] | Teixobactin was isolated from a new species, <i>Eleftheria terrae</i>, from soil using a [http://en.wikipedia.org/wiki/High-throughput_screening high-throughput] screening device called the iChip.[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>]The iChip, a collection of plates with hundreds of wells covered by two layers of semi-permeable membranes, is used to isolate previously unculturable bacteria by growing microbes placed in the device [http://en.wikipedia.org/wiki/In_situ <i>in situ</i>]where their typical environmental factors are used to cultivate the bacteria [http://aem.asm.org/content/76/8/2445.full <sup>4</sup>] In this case, a soil sample was diluted so that on average one bacteria cell could be introduced into each iChip well, and the device was covered by the semi-permeable membranes to be placed back into the soil where the bacteria was originally collected.[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>] Compared to standard Petri dishes, the colony count for soil bacteria using the iChip was 5 times higher when cultivating from a single colony's worth of bacteria.[http://aem.asm.org/content/76/8/2445.full <sup>4</sup>] Extracts from thousands of isolates were tested for antibiotic activity, and the discovery of teixobactin's properties lead to the identification and classification of <i>E. terrae</i> using [http://en.wikipedia.org/wiki/16S_ribosomal_RNA 16S rRNA gene] sequencing.[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>] Relatives of <i>E. terrae</i> were not previously known to produce antibiotics.[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>] Though the use of the iChip's cultivating abilities, other antibiotics like teixobactin could be discovered. | ||
==Mechanism of Action, | ==Mechanism of Action, Efficacy, and Resistance of Teixobactin== | ||
[[Image: TeixobactinMIC.png |thumb|450px|right|A comparison of Teixobactin's activity against gram negative and gram positive bacteria using the MIC data determined by Ling <i>et al</i> via [http://en.wikipedia.org/wiki/Broth_microdilution broth microdilution].[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>] Teixobactin was extremely effective against gram positive bacteria, but not gram negative bacteria where the outer membrane prevents the drug from binding to lipid precursors in the cell wall layer.[http://www.nature.com/nature/journal/v517/n7535/full/nature14193.html <sup> | [[Image:TeixobactinMIC.png |thumb|450px|right|A comparison of Teixobactin's activity against gram negative and gram positive bacteria using the MIC data determined by Ling <i>et al</i> via [http://en.wikipedia.org/wiki/Broth_microdilution broth microdilution].[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>] Teixobactin was extremely effective against gram positive bacteria, but not gram negative bacteria where the outer membrane prevents the drug from binding to lipid precursors in the cell wall layer.[http://www.nature.com/nature/journal/v517/n7535/full/nature14193.html <sup>2</sup>] Image created by Lisette Espinosa.]] | ||
[http://en.wikipedia.org/wiki/Minimum_inhibitory_concentration Minimum inhibitory concentrations] (MIC) were used to measure the effectiveness of teixobactin against pathogenic bacteria.[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>] While teixobactin was not effective against gram-negative bacteria, less than 0.6 μg/mL of the drug was necessary to visibly inhibit the growth of many gram positive bacteria tested including pathogenic MRSA, VRE, Bacillus anthracis, and Clostridium difficile.[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>] No toxicity toward mammalian cells was found as expected from the [http://en.wikipedia.org/wiki/Mechanism_of_action mechanism of action] . The antibiotic works through inhibiting cell wall synthesis by binding to two cell wall precursors, lipid II and lipid III. [http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>] Though researchers looked for possible teixobactin-resistant mutants using S. aureau and M. tuberculosis through plating the bacteria in media with four times their respective MICs, no resistant mutants were found.[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>] Studies done in the past on vancomycin, another antibiotic that binds to lipid cell wall precursors, suggest that teixobactin's lack of development of resistance may be due to the fact that the lipids are made from organic precursors rather than synthesized [http://en.wikipedia.org/wiki/De_novo de novo] from DNA like proteins, making development of resistance through mutation alone more difficult.[http://www.nature.com/nature/journal/v517/n7535/full/nature14193.html <sup> | [[Image:TeixobactinMOA.png|thumb|600px|right|Simplified schematic of the inhibition of cell wall synthesis by teixobactin in a gram positive bacterium. Teixobactin inhibits both teichoic acid and peptidoglycan synthesis in gram positive cells. "Gram-positive cell wall schematic" by Twooars at the English language Wikipedia.[http://commons.wikimedia.org/wiki/File:Gram-positive_cellwall-schematic.png#/media/File:Gram-positive_cellwall-schematic.png] Modified by Lisette Espinosa. | ||
]] | |||
[http://en.wikipedia.org/wiki/Minimum_inhibitory_concentration Minimum inhibitory concentrations] (MIC) were used to measure the effectiveness of teixobactin against pathogenic bacteria.[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>] While teixobactin was not effective against gram-negative bacteria, less than 0.6 μg/mL of the drug was necessary to visibly inhibit the growth of many gram positive bacteria tested including pathogenic MRSA, VRE, <i>Bacillus anthracis</i>, and <i>Clostridium difficile</i>.[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>] No toxicity toward mammalian cells was found as expected from the [http://en.wikipedia.org/wiki/Mechanism_of_action mechanism of action]. The antibiotic works through inhibiting cell wall synthesis by binding to two cell wall precursors, lipid II and lipid III. Lipid II is precursor for [http://en.wikipedia.org/wiki/Peptidoglycan peptidoglycan], a polymer that forms the cell wall, and lipid III is a precursor for [http://en.wikipedia.org/wiki/Teichoic_acid techoic acid], polymers that provide rigidity to the cell wall in gram positive bacteria.[http://www.nature.com/nature/journal/v517/n7535/full/nature14193.html <sup>2</sup>] The specificity of teixobactin to the lipid cell wall precursors and the presence of an outer cell membrane is gram negative cells explain the antibiotic's lack of effectiveness on gram negative cells. [http://www.nature.com/nature/journal/v517/n7535/full/nature14193.html <sup>2</sup>] Teixobactin blocks precursor recycling as well as techoic acid and peptidoglycan synthesis. | |||
[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>] Though researchers looked for possible teixobactin-resistant mutants using <i>S. aureau</i> and <i>M. tuberculosis</i> through plating the bacteria in media with four times their respective MICs, no resistant mutants were found.[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>] Studies done in the past on vancomycin, another antibiotic that binds to lipid cell wall precursors, suggest that teixobactin's lack of development of resistance may be due to the fact that the lipids are made from organic precursors rather than synthesized [http://en.wikipedia.org/wiki/De_novo de novo] from DNA like proteins, making development of resistance through mutation alone more difficult.[http://www.nature.com/nature/journal/v517/n7535/full/nature14193.html <sup>2</sup>] In addition, the binding target of the drug is highly conserved among bacteria.[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>] Resistance to vancomycin was identified almost 40 years after the drug's discovery when it is believed that the self-resistance vector from vanomycin-producing bacteria was captured by pathogenic bacteria through [http://en.wikipedia.org/wiki/Horizontal_gene_transfer horizontal gene transfer].[http://www.nature.com/nature/journal/v517/n7535/full/nature14193.html <sup>2</sup>] | |||
== Limitations of Teixobactin== | == Limitations of Teixobactin== | ||
Although resistance to teixobactin was difficult to manufacture in lab, resistance could eventually emerge in the same manner vancomycin resistance emerged, through horizontal gene transfer.[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>] However, as <i>E. terrae</i> is gram negative and does not need carry genes for self-resistance like vancomycin-producing bacteria, the genes for resistance would likely come from other soil bacteria.[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>] | Although resistance to teixobactin was difficult to manufacture in lab, resistance could eventually emerge in the same manner vancomycin resistance emerged, through horizontal gene transfer.[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>] However, as <i>E. terrae</i> is gram negative and does not need carry genes for self-resistance like vancomycin-producing bacteria, the genes for resistance would likely come from other soil bacteria.[http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html <sup>1</sup>] As teixobactin is a new drug, clinical trials on humans have not yet been conducted. Kim Lewis, a co-researcher in the Ling <i>et al.</i> study, speculated that teixobactin could be in trial by 2017.[http://cen.acs.org/articles/93/web/2015/01/Novel-Bacteria-Fighter-Unearthed.html <sup>5</sup>] | ||
==References== | ==References== | ||
1. [http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html Ling, L., Schneider, T., Peoples, A., Spoering, A., Engels, I., & Conlon, B. et al. (2015). A new antibiotic kills pathogens without detectable resistance. Nature, 517(7535), 455-459. doi:10.1038/nature14098] | 1. [http://www.nature.com/nature/journal/v517/n7535/full/nature14098.html Ling, L., Schneider, T., Peoples, A., Spoering, A., Engels, I., & Conlon, B. et al. (2015). A new antibiotic kills pathogens without detectable resistance. Nature, 517(7535), 455-459. doi:10.1038/nature14098] | ||
2. [http:// | 2. [http://www.nature.com/nature/journal/v517/n7535/full/nature14193.html Wright, G. (2015). Antibiotics: An irresistible newcomer. Nature, 517(7535), 442-444. doi:10.1038/nature14193] | ||
3. [http://static.squarespace.com/static/54878590e4b0b8980d215324/t/54ad7aaae4b09209a6148464/1420655274323/2015+january+7th+Novobiotic+press+release.pdf Lewis, Kim (7 January 2015). NovoBiotic reports the discovery of teixobactin, a new antibiotic without detectable resistance. Cambridge, Massachusetts: NovoBiotic Pharmaceuticals. Retrieved 7 January 2015] | 3. [http://static.squarespace.com/static/54878590e4b0b8980d215324/t/54ad7aaae4b09209a6148464/1420655274323/2015+january+7th+Novobiotic+press+release.pdf Lewis, Kim (7 January 2015). NovoBiotic reports the discovery of teixobactin, a new antibiotic without detectable resistance. Cambridge, Massachusetts: NovoBiotic Pharmaceuticals. Retrieved 7 January 2015] | ||
4. [http:// | 4. [http://aem.asm.org/content/76/8/2445.full Nichols, D., Cahoon, N., Trakhtenberg, E., Pham, L., Mehta, A., & Belanger, A. et al. (2010). Use of Ichip for High-Throughput In Situ Cultivation of "Uncultivable" Microbial Species. Applied And Environmental Microbiology, 76(8), 2445-2450. doi:10.1128/aem.01754-09] | ||
5. [http://cen.acs.org/articles/93/web/2015/01/Novel-Bacteria-Fighter-Unearthed.html Halford, B. (2015). Novel Bacteria-Fighter Unearthed. Chemical & Engineering News. Cen.acs.org. Retrieved 24 March 2015.] | 5. [http://cen.acs.org/articles/93/web/2015/01/Novel-Bacteria-Fighter-Unearthed.html Halford, B. (2015). Novel Bacteria-Fighter Unearthed. Chemical & Engineering News. Cen.acs.org. Retrieved 24 March 2015.] | ||
Latest revision as of 23:48, 14 April 2015
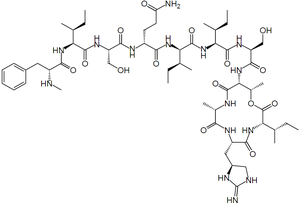
The development of new antibiotics has been a difficult task further complicated by the the rapid evolution of resistant bacteria. Teixobactin is a newly discovered antibiotic that is effective against gram positive bacteria including antibiotic-resistant strains without evidence of resistance development.1 The antibiotic, isolated from the soil using a device called the iChip, works by inhibiting cell wall synthesis.1
Introduction
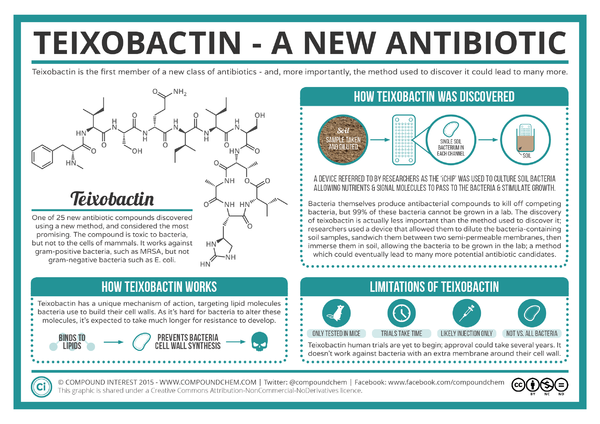
Microorganisms have been our primary source of new antibiotics since the drugs' introduction in the 1940s.1 However, only about 1% of all bacterial species are readily culturable in laboratory conditions.1 Despite the need for new drugs, finding new antibiotics is expensive and difficult, and the identification of new, effective compounds have dramatically decreased since the mid-twentieth century.2 NovoBiotic Pharmaceuticals in collaboration with the University of Bonn and Northeastern University reported in Nature the discovery of a new antibiotic, teixobactin, with properties that minimize the development of bacterial resistance.3 Ling et al. found teixobactin to be effective against gram positive bacteria, including drug-resistant strains such as methicilin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE).1 This antibiotic, isolated using a device called the iChip, works through inhibiting cell wall synthesis by binding to two lipid cell wall precursors, lipid II and lipid III.1 Teixobactin was highly effective in treating MRSA and Streptococcus pneumoniae in mice via injection.1
Identification
Teixobactin was isolated from a new species, Eleftheria terrae, from soil using a high-throughput screening device called the iChip.1The iChip, a collection of plates with hundreds of wells covered by two layers of semi-permeable membranes, is used to isolate previously unculturable bacteria by growing microbes placed in the device in situwhere their typical environmental factors are used to cultivate the bacteria 4 In this case, a soil sample was diluted so that on average one bacteria cell could be introduced into each iChip well, and the device was covered by the semi-permeable membranes to be placed back into the soil where the bacteria was originally collected.1 Compared to standard Petri dishes, the colony count for soil bacteria using the iChip was 5 times higher when cultivating from a single colony's worth of bacteria.4 Extracts from thousands of isolates were tested for antibiotic activity, and the discovery of teixobactin's properties lead to the identification and classification of E. terrae using 16S rRNA gene sequencing.1 Relatives of E. terrae were not previously known to produce antibiotics.1 Though the use of the iChip's cultivating abilities, other antibiotics like teixobactin could be discovered.
Mechanism of Action, Efficacy, and Resistance of Teixobactin
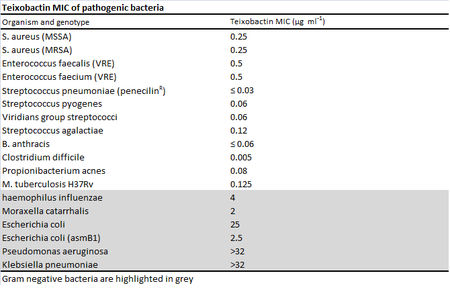
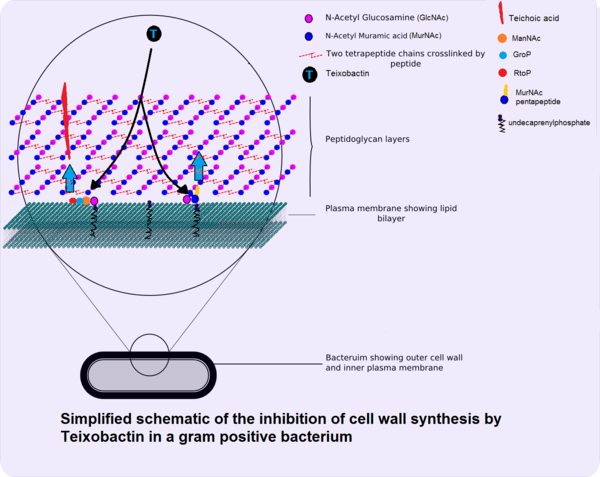
Minimum inhibitory concentrations (MIC) were used to measure the effectiveness of teixobactin against pathogenic bacteria.1 While teixobactin was not effective against gram-negative bacteria, less than 0.6 μg/mL of the drug was necessary to visibly inhibit the growth of many gram positive bacteria tested including pathogenic MRSA, VRE, Bacillus anthracis, and Clostridium difficile.1 No toxicity toward mammalian cells was found as expected from the mechanism of action. The antibiotic works through inhibiting cell wall synthesis by binding to two cell wall precursors, lipid II and lipid III. Lipid II is precursor for peptidoglycan, a polymer that forms the cell wall, and lipid III is a precursor for techoic acid, polymers that provide rigidity to the cell wall in gram positive bacteria.2 The specificity of teixobactin to the lipid cell wall precursors and the presence of an outer cell membrane is gram negative cells explain the antibiotic's lack of effectiveness on gram negative cells. 2 Teixobactin blocks precursor recycling as well as techoic acid and peptidoglycan synthesis.
1 Though researchers looked for possible teixobactin-resistant mutants using S. aureau and M. tuberculosis through plating the bacteria in media with four times their respective MICs, no resistant mutants were found.1 Studies done in the past on vancomycin, another antibiotic that binds to lipid cell wall precursors, suggest that teixobactin's lack of development of resistance may be due to the fact that the lipids are made from organic precursors rather than synthesized de novo from DNA like proteins, making development of resistance through mutation alone more difficult.2 In addition, the binding target of the drug is highly conserved among bacteria.1 Resistance to vancomycin was identified almost 40 years after the drug's discovery when it is believed that the self-resistance vector from vanomycin-producing bacteria was captured by pathogenic bacteria through horizontal gene transfer.2
Limitations of Teixobactin
Although resistance to teixobactin was difficult to manufacture in lab, resistance could eventually emerge in the same manner vancomycin resistance emerged, through horizontal gene transfer.1 However, as E. terrae is gram negative and does not need carry genes for self-resistance like vancomycin-producing bacteria, the genes for resistance would likely come from other soil bacteria.1 As teixobactin is a new drug, clinical trials on humans have not yet been conducted. Kim Lewis, a co-researcher in the Ling et al. study, speculated that teixobactin could be in trial by 2017.5
References
Edited by Lisette Espinosa, a student of Nora Sullivan in BIOL168L (Microbiology) in The Keck Science Department of the Claremont Colleges Spring 2014.
