Shigellosis: Difference between revisions
No edit summary |
No edit summary |
||
| (One intermediate revision by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{ | {{Conway}} | ||
[[Image:ShigellaOnHe-La.jpeg|thumb|400px|right|''Shigella'' bacteria, in red, shown on the surface of an He-La cell. From: http://www.cimst.ethz.ch/ [http://www.cimst.ethz.ch/gallery]]] | [[Image:ShigellaOnHe-La.jpeg|thumb|400px|right|''Shigella'' bacteria, in red, shown on the surface of an He-La cell. From: http://www.cimst.ethz.ch/ [http://www.cimst.ethz.ch/gallery]]] | ||
==Etiology/Bacteriology== | ==Etiology/Bacteriology== | ||
| Line 6: | Line 5: | ||
| Domain = [[Bacteria]] | | Domain = [[Bacteria]] | ||
| Phylum = [[Proteobacteria]] | | Phylum = [[Proteobacteria]] | ||
| Class = | | Class = Gammaproteobacteria | ||
| Order = [[Enterobacteriales]] | | Order = [[Enterobacteriales]] | ||
| Family = Enterobacteriaceae | | Family = Enterobacteriaceae | ||
Latest revision as of 16:45, 11 February 2016

Etiology/Bacteriology
Taxonomy
| Domain = Bacteria | Phylum = Proteobacteria | Class = Gammaproteobacteria | Order = Enterobacteriales | Family = Enterobacteriaceae | Genus = Shigella
Description
Shigellosis is caused by a bacterial infection of the Shigella genus. Shigella bacteria are Gram-negative, rod-shaped, non spore-forming, facultative anaerobes. There are four Shigella species, only three of which are the major disease causing species. S. flexneri accounts for roughly 60% of shigellosis cases in the developing world and is the most frequently isolated species of Shigella worldwide [1]. S. sonnei is the cause of 77% of cases in the developed world while only causing 15% of cases in the developing world, and it is also causes two-thirds of shigellosis cases in the United States [1]. S. dysenteriae is the most common cause of dysentery epidemics, usually in confined populations with poor hygiene and little to no sanitation, for example environments such as refugee camps. S. dysenteriae is a major concern because it is the only species of Shigella that produces the Shiga toxin.
Pathogenesis
Virulence factors
Shigella species pose a wide variety of virulence factors that allow it to adhere to the epithelium of the intestine, survive stomach acid, invade host cells, evade immune responses, and introduce toxins into the body.
Acid tolerance
Shigella’s ability to tolerate acid contributes to its low infectious dose, however, research has shown that acid tolerance changes according to growth phase [2]. Stationary phase seems to be necessary in order for the pathogen to survive conditions of low pH.
Effector proteins
Several effector proteins encoded on Shigella’s virulence plasmid allow the pathogen to invade host cells and move through the eukaryotic cytoplasm. Many of these same proteins help the pathogen adhere to the host cell wall. Invasion of plasmid antigen B (IpaB) initiates binding to the host cell while also initiating pathways that kill macrophages upon infection, IpaC activates proteins to form the actin-polymerizing complex that allows Shigella to move and spread within host cells [3].
The Mxi-Spa T3SS
Shigella utilizes a Type III Secretion System to deliver its effector protein to the cytoplasm of host cells. Transmission electron microscopy reveals the ATPase- dependent T3SS as a 45 to 60 nm needle that extends out from the bacterium nested in a complex composed of seven rings primarily made of MxiD [4]. The whole complex spans the inner membrane, periplasm, and outer membrane of the pathogen and continues to allow for binding to host cells. As the internal of the diameter of the needle is only 2 to 3 nm across, effector proteins pass through the complex in a denatured form.
In order to control and time secretion of effector proteins, Shigella senses environmental oxygen conditions and utilizes essential effector proteins. In low oxygen environments, the pathogen elongates the T3SS needle thus preventing effector proteins from being secreted. As the bacterium approaches host cells, which require oxygen, it glycosylates its LPS in order to shorten them and thus expose the T3SS needle [5]. Once Shigella has translocated IpaB and IpaC to the tip of the needle, IpaB binds to the host cell surface, enduces changes in the cell membrane of the host cell, and works with IpaC to insert the needle into the host cell.
Toxins
Three toxins are principally associated with Shigella species and account for, or are hypothesized to account, for the emergence of bloody diarrhea in infected humans. Shigella enterotoxin 1 and 2 (ShET1 and ShET2), produced by the bacteria Shigella dysenteriae and some strains of Escherichia coli, were shown to induce fluid secretion in the intestine and thus thought to produce the watery part of diarrhea. However, the exact mechanism that results in diarrhea is still unknown [6].
Shiga toxin, on the other hand, is only associated with S. dysenteriae and accounts for the life-threatening symptoms found in some Shigella infections. Shiga toxin is an AB toxin consisting of a ~32 kDa A subunit and a pentamer of ~7 kDa B subunits. Once released from the bacterium, the B subunits bind globotriaosylceramide on the surface of the host cell. The toxin is then internalized and moved toward the cell by the Golgi network through a process called retrograde transport [6]. During this process, the A subunit is cleaved to created a A1 fragment with N- glycosidase activity, which cleaves an adenine from the 28s rRNA component of the 60s ribosomal subunit once the toxin reaches the endoplasmic reticulum. The cleavage inhibits EF1 mediated tRNA bonding and halts elongation of peptides, inhibiting protein synthesis.
Mechanism
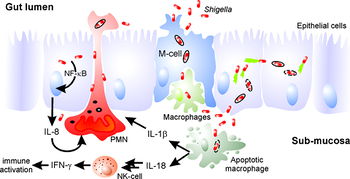
The pathogenesis of "Shigella" revolves around the bacterium's ability to enter host cells and hijack cellular machinery in order to evade immune responses and renew its replicative space. By utilizing an injectisome, the pathogen can inject effector proteins into the host cell that allow it to manipulate the eukaryotic cell's cytoskeleton, primarily actin fibers. Actin polymerization drives the process that leads to uptake of the pathogen and gives the bacterium a way to achieve intracellular motility as well as a way to pass laterally to other host cells. In addition, Shigella can mediate the cell death of macrophages to further evade the host's immune system.
Adherence
Once Shigella spp passes through the stomach, the pathogen approaches the large intestine where it will eventually cross the intestinal epithelial through a microfold cell to induce its pathogenicity. M cells, located in the Peyer’s patch, facilitate the development of immune response by absorbing antigens and passing them on to lymphoid tissues, and Shigella utilizes this function to gain access to the basolateral side of the epithelial cells.
Shigella adheres to M cells by binding CD44 and α5β1 found in areas of cell membrane high in cholesterol content known as lipid rafts [7]. These rafts are known to contain bunches of specific proteins, and in this case, the rafts contain the aforementioned receptors recognized by IpaB and the IpaBCD effector proteins on the pathogen’s surface. Binding initiates the introduction of the T3SS needle into the cell membrane which initiates the rearrangement of the membrane to bring more lipid rafts to the bacterial entry site and thus bringing more receptors to the cell and creating a signaling platform [7].
Entering the cell
Successful adherence prompts the pathogen to secrete various effector proteins through its T3SS that will help force the cell to ingest the bacterium by rearranging the eukaryotic cytoskeleton. IpgD and IpaA work to break down existing actin polymers and disconnect the cytoskeleton from the cytoplasmic side of the membrane respectively. VirA destabilizes microtubules and simultaneously activates a GTPase called Rac1. VirA works alongside IpaC and IpgB2 to activate Rac1 and Cdc42, another GTPase, which recruit Arp2/3 to initiate actin nucleation [8]. The following actin polymerization creates protrusions that extend from the host cell and then engulf the pathogen thus enclosing Shigella in a phagosome and giving it access to the inside of the cell.
Having passed into the M cell, Shigella utilizes its effector proteins and T3SS to escape from the phagosome—a process which occurs within the span of 15 minutes [9]
. Shigella begins to multiply inside of the host cell after breaking down the phagosome and secretes IscB, which competitively binds to IscA, to avoid autophagy in the cell. In addition, IpgD actives signal pathways that prevent the host cell from inducing apoptosis in order the pathogen to multiply without interruption.
Actin-mediated motility
Movement within the cytoplasm of the cell is achieved by hijacking actin-nucleating machinery. Surface-bound IscA binds and activates N-WASP, which recruits Arp2/3. Polymerization of actin on one end of the pathogen pushes it through the cytoplasm in the opposite direction, and VirA destabilizes microtubules surrounding the pathogen in order to allow movement through the cytoskeleton [10]. This induced motility allows the pathogen to evade extracellular immune defenses and invade neighboring epithelial cells by pressing against the cell membrane to create a protrusion that is endocytosis by the neighboring cell where it will multiply and continue to infect the epithelium [11]. Shigella can use the same mechanism to break through the basolateral cell membrane and reach the intestinal mucosa. The spread of Shigella in the epithelium through this process results in pronounced death of epithelial cells and subsequently a strong immune response.
Encountering macrophages
Shigella encounters macrophages shortly after reaching the intestinal mucosa. Shigella breaks through the phagosome upon being phagocytized and begins to secrete IpaB in the cytoplasm of the macrophage. IpaB binds to caspase-1, a protease in the macrophage, to induce pyroptosis through a variety of pathways including DNA fragmentation, ATP loss and cell membrane rupture [12]. The death of the macrophage triggers a release of cytokines that play a role in the massive inflammatory response associated with Shigella infection [3]. This ability to invade and destroy macrophages allows the pathogen to evade a key part of the host’s innate immune response and constitutes a key part of its pathogenesis.
Clinical features
Symptoms commonly set in 1 to 2 days after being exposed to the bacteria and include bloody diarrhea, fever, and stomach cramps. Shigellosis typically passes within five to seven days. The Shiga toxin produced by S. dysenteriae can cause Hemolytic-uremic syndrome (HUS), and symptoms include bloody urine, kidney failure, and low blood platelet count, which help to clot blood [13]. These symptoms usually set in five to ten days after diarrheal manifestation.
Diagnosis
Observation of bloody diarrhea alone cannot be used to determine Shigella to be the causative agent. Stool samples from the patient are needed to culture bacteria and confirm the presence of a Shigella species. The presence of leukocytes in the stool also signifies Shigella as the probable causative agent [13]. Approximately 70 percent of patients suffering from shigellosis have leukocytes or fecal blood detected in the stool sample.
Treatment
Mild cases of Shigellosis can be managed without antibiotics, and recovery can be fairly quick. Antibiotics can limit the spread of the disease in the intestine and reduce the time it takes for the disease to pass through the body. For immunocompromised patients or patients demonstrating severe complications, antibiotics can be necessary. Susceptibility tests should be run to see if the bacterium is resistant to drugs like ampicillin. If so, antibiotics like flouroquinolones and azithromycin may be needed. Typically, the use of antidiarrheal medicines is not recommended because it may worsen the disease [15]. The use of bacteriophages is another common method for treating Shigellosis. Phages for treating Shigellosis have been isolated from raw sewage, environmental waters, soil, farm and food processing effluents, manure, retail foods and foods of dairy origin. Technological developments have recently been made in phage therapy as constant improvement of rapid and cost effective genome sequencing technologies ensures that all phages used in phage therapy approaches will be properly assessed for safety [16].
Vaccination
Currently, there is no licensed vaccine available targeting Shigella. Vaccination is difficult for Shigellosis because it is based on a narrow, type-specific O-somatic antigen. However, there are multiple research groups that working on vaccines using very unique approaches. One group passively administered Immunoglobulin from cows milk with a High-titer of S. flexneri antibodies which resulted in protection from wild type infection of S. flexneri. Yet, the administration of low titer antibodies in cow's milk did not yield protection. Also, another group has made progress through live oral vaccinations, but a difficult problem they are facing is that there is a small margin between under and over attenuation [15]. Children seem to develop under attenuation to the vaccine which causes excessive reactogenicity. Developing countries have the other problem of over attenuation to the live oral vaccinations which doesn't provide sufficient immmunogenicity. Whole cell vaccines are being tested as vaccinations at John's Hopkins using S.sonnei and at Antex using Shigella. Clinical testing of these two vaccines is near. Vaccines are being tested on pigs where the toxins of the Shigella species are placed on other bacteria, results have yielded protection.
Prevention
No effective vaccine for shigellosis currently exists, but person-to-person trasmission can be prevented through frequent, thorough hand washing [17]. Children should also practice good hand washing to prevent infection obtained from a daycare center or through improper toilet use. Special care should be taken when a child in diapers presents shigellosis. Diapers should be disposed in closed-lid waste bins, and whoever handled the diaper should wash his or her hands as well as the child’s hands once the changing is over. The changing area should also be thoroughly cleaned with bactericidal agents. Child-to-child contact should be avoided if possible if a child has shigellosis.
People with shigellosis should not prepare food for others until they have proven that they are no longer infected. Typical food and water safety precautions should be sufficient to prevent the disease. Greater food safety should be taken when in the developing world. Food should only be consumed if you have treated, cooked, or peeled it for yourself, and water should be treated or boiled before consumption [16].
Host Immune Response
Massive inflammatory response, a principal feature of shigellosis, is induced by the pathogen through a variety of pathways. Epithelial cells possess pathogen recognition receptors that trigger signaling pathways once Shigella enters the cytoplasm, and macrophages induce similar responses upon being invaded by the pathogen. In the face of the immune response, Shigella employs strategies that allow the bacterium to downregulate certain pathways and avoid intricate defense mechanisms.
Inside epithelial cells
Peptidoglycan molecules released by Shigella are recognized by nucleotide-binding oligomerization domain-containing protein 1 (NOD1) once the bacterium enters the cytoplasm, and a signal transduction event follows that activates MAPK and NF-kB pathways to initiate the secretion of cytokines and antimicrobial peptides which trigger an inflammatory response [5]. These signaling molecules travel to neighboring cells through connexin gap junctions thus activating inflammatory response pathways in these cells. Since Shigella can travel between cells without leaving the cytoplasm, this universal response helps prep the epithelial lining for further invasion.
Autophagic recognition
Autophagy protein 5(Atg5) recognizes and binds VirG, an effector protein that helps activate the actin polymerization needed for Shigella to achieve intracellular motility, and to initiate autophagosome formation. Binding of Atg5 to VirG also induces a response that causes proteins called septins to accumulate at bacterial entry foci and form cages that trap the invading bacterium [5]. Once incased in the septin cage, the pathogen cannot move throughout the cell and subsequently destroy by autophagic pathways.
Effector protein IcsB, however, competitively binds to VirG to prevent recognition by Atg5. Without an Atg5-mediated response, Shigella is able to move and multiply in the cytoplasm out of grasp of autophagy [3]. Such avoidance strategies help illustrate the pathogen’s detailed pathogenesis.
Recruitment of leukocytes
Chemokines released by infected epithelial cells and macrophages attract leukocytes to the site of infection—one of the first steps toward resolving the infection. Apart from phagocytizing and killing the pathogen, neutrophils specifically utilize a proteinase that preferentially degrades Shigella effector proteins and produces neutrophil extracellular traps that catch extracellular pathogens and further degrade effector proteins [5].
Extravasation of neutrophils, however, initially makes the infection worse by disrupting cell-to-cell junctions that would otherwise limit Shigella's access to the basolateral surface of epithelial tissue [5].
In macrophages
While the invasion and destruction of macrophages is a key part of Shigella pathogenesis, the leukocyte still induces responses that predispose the body to counteracting the infection. Macrophages mediate an immune response through the secretion of cytokines IL-1beta and IL-18, and eventual death of the macrophage also releases molecules that help the mucosal immune system respond to the infection [5]. A notable response is the secretion of IgA, which helps neutralize the pathogen and prevents its adherence, and IgG, which specifically targets LPS on the Gram-negative pathogen. This is a key part that helps prevent infection with the same serotype of pathogen.
References
1 World Health Organization Initiative for Vaccine Research. Shigellosis overview
14 M.S. Parmar. Medscape Reference. Hemolytic-Uremic Syndrome. Apr 18, 2013.
15 J.A. Kroser. Medscape Reference. Shigellosis Workup. May 23, 2013.
17 J. Sureshbabu. Medscape Reference. Shigella Infection Workup. Jun 25, 2012.
Created by Jake Morgan, Michael McCoy and Kyle Trinidad, students of Tyrrell Conway at the University of Oklahoma.
