SungTimon2.2: Difference between revisions
Kaleen.timon (talk | contribs) |
Kaleen.timon (talk | contribs) |
||
| (127 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
==Classification== | ==Classification== | ||
Kingdom- Bacteria; Phylum- Firmicutes; Class- Bacilli; Order- Bacillales; Family- Bacillaceae; Genus- Bacillus; Species- Bacillus Megaterium | |||
Our Blast results were inconclusive. Bacillus Megaterium is the "best guess" narrowed down from our biochemical results and cellular and colonial characteristics in the lab** | |||
===Species=== | ===Species=== | ||
''Genus species'' | |||
'' | Bacillus Megaterium | ||
(Our second choice was Bacillus Cereus) | |||
==Habitat Information == | ==Habitat Information == | ||
The soil organism was isolated from a partially shaded creek at the San Marcos Golf Course in San Marcos, Tx on 09/02/15. The air temperature was 90F/32.2C, humidity 49%, pressure 30.04, solar radiation UV N/A. The grid coordinates from NRCS are 29 53.6' 97 55.4'. Spores are known to occur in soil, dust, water and plants. | |||
==Description and Significance== | ==Description and Significance== | ||
This organism is a gram positive, endospore forming, rod shaped bacteria. The colonial morphology is circular, smooth in the center, 'grainy' on the outside, undulate margins, umbonate elevation, and opaque with a golden appearance. The cellular arrangement is bacillus. | |||
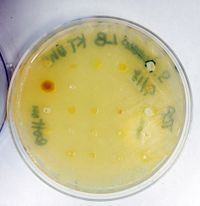
[[File:116.jpeg|200px|thumb|left|Kirby Bauer]] | |||
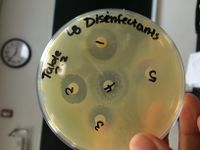
[[File:117.jpeg|200px|thumb|left|Disinfectant Test]] | |||
Kirby Bauer Antimicrobial | |||
Strong Zone of Inhibition for linezolid, cefamandole, azlocillin, vancomycin, sufisozazole | |||
Weak Zone of Inhibition for | |||
Kirby Bauer Antimicrobial results: | |||
Strong Zone of Inhibition for (1) linezolid, (2) cefamandole, (3) azlocillin, (4) vancomycin, (5) sufisozazole. | |||
Weak Zone of Inhibition for (6) oxacillin. | |||
Disinfectant Sensitivity Results: | |||
Strong Zone of Inhibition for (1) clove, (2) 10% lysol. | |||
Weak Zone of Inhibition for (3) tea tree oil, (4) 100% bleach, and (5) lavender. | |||
Anti-microbial activity was observed on an S. aureus test patch plate. | |||
[[File:118.jpeg|200px|thumb|left|S. aureus lawn patch plate]] | |||
B. megaterium is a significant organism because it can be used industrially as a commercial strain and expression host [1]. B. megaterium has important implications as a host for penicillin aminidase and vitamin B12 [3]. A major advantage is that B. megaterium can secrete proteins directly into the growth medium [6]. Some strains are capable of nitrogen fixation [2]. | |||
==Genome Structure== | ==Genome Structure== | ||
The size and content of the genome are undetermined due to inconclusive Blast results. PCR and DNA sequencing did not yield an adequate sequence for organism identification. Therefore, our genome information is based off of a 'best guess' of what organism might be based on biochemical and staining results. S Ribosomal sequence for our soil organism is not available. | |||
The industrially significant B. megaterium strain QMB1551 has a 5.1 Mbp chromosome that carries approximately 5,300 genes. QMB1551 contains 7 natural plasmid (spBM100 to pBM700) ranging from 5.4 kb to over 164 kb. The chromosome is circular with an average G/C content of 38.2%. In 2011, Eppinger, et. al. completed the genomic sequence of QMB1551 [3]. | |||
==Cell Structure, Metabolism and Life Cycle== | |||
The vegetative surface of B. megaterium consists of a proteinaceous surface layer (S-layer) and a cell wall made of peptidiglycan, containing meso-diaminopimelic acid (DAP). The function of the proteinaceous surface layer, made of crystalline glycoprotein subunits, is unknown. It may be involved in bacterial metal interactions [4]. Capsules may be present but were not observed for our soil organism. B. megaterium is mobile via a peritrichous flagella. | |||
B. megaterium utilizes aerobic metabolism but has been known to survive anaerobic conditions. B. megaterium can produce metabolically inactive endospores to survive harsh conditions. The life cycle involves the following [5]: | |||
1) Vegetative cell | |||
2) Vegetative forms a septum (1/3 of the length of the cell) and pumps DNA into septum | |||
3) Endospore develops and then surrounded by keratin-like coat | |||
4) Vegetative cell dies; endospore release | |||
5) Free mature endospore | |||
6) Endospore becomes a vegetative cell | |||
This is a link to endospore staining of B. megaterium: | |||
http://www.microbelibrary.org/images/tu/bac_meg_endosp1.jpg | |||
==Physiology and Pathogenesis== | ==Physiology and Pathogenesis== | ||
B. megaterium is a common bacteria found in soil, does not produce endotoxins, and is non-pathogenic. | |||
[[File:4275.jpeg|200px|thumb|left|Phenol Red]] | |||
[[File:111.jpeg|200px|thumb|left|DNAse]] | |||
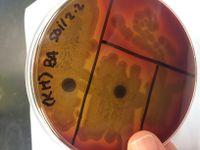
[[File:112.jpeg|200px|thumb|left|Blood Agar]] | |||
[[File:113.jpeg|200px|thumb|left|PEA]] | |||
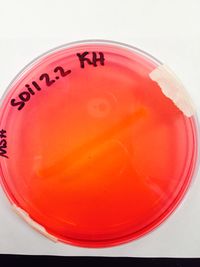
[[File:114.jpeg|200px|thumb|left|Mannitol Salt Agar]] | |||
[[File:115.jpeg|200px|thumb|left|Lipid Hydrolysis]] | |||
NEGATIVE RESULTS INCLUDE: Starch Hydrolysis, Gelatin Hydrolysis, Methyl Red-Voges Proskauer, Citrate test, SIM (negative for sulfur reduction, indole production, nonmotile organism), nitrate reduction, Urea hydrolysis, Eosin methylene blue (no coliforms, lactose fermenters, no growth: meaning possible gram +), Hektoen Enteric (presumptive ID for Gram +), MacConkey Agar (Gram + evidence), Decarboxylation Test, Phenylalanine deaminase, and | |||
Biochemical characteristics: | |||
NEGATIVE RESULTS INCLUDE: *Starch Hydrolysis, *Gelatin Hydrolysis, Methyl Red-Voges Proskauer, *Citrate test, SIM (negative for sulfur reduction, indole production, nonmotile organism), nitrate reduction, Urea hydrolysis, Eosin methylene blue (no coliforms, lactose fermenters, no growth: meaning possible gram +), Hektoen Enteric (presumptive ID for Gram +), MacConkey Agar (Gram + evidence), Decarboxylation Test, Phenylalanine deaminase, and *Catalase. | |||
POSITIVE RESULTS INCLUDE: Casein hydrolysis, DNA hydrolysis, Lipid hydrolysis, Triple sugar iron, oxidase, blood agar (beta- complete lysis of RBC), mannitol salt agar (weak positive), phenylethal alcohol, and bile esculin. | POSITIVE RESULTS INCLUDE: Casein hydrolysis, DNA hydrolysis, Lipid hydrolysis, Triple sugar iron, oxidase, blood agar (beta- complete lysis of RBC), mannitol salt agar (weak positive), phenylethal alcohol, and bile esculin. | ||
Interpretation of Results: Our bacteria is a gram positive organism that possesses the enzymes casease,DNase, ligase, fermentable enzymes, and cytochrome C oxidase. This organism can completely lyse red blood cells, and can weakly ferment mannitol. | Interpretation of Results: Our bacteria is a gram positive organism that possesses the enzymes casease, DNase, ligase, fermentable enzymes, and cytochrome C oxidase. This organism can completely lyse red blood cells, and can weakly ferment mannitol. | ||
* *indicates inconsistent result with literature, likely due to "best guess" method. | |||
==References== | ==References== | ||
[ | http://www.eol.org/pages/974186/overview | ||
[1] [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1392972/ Malten, M., Biedendieck, R., Gamer, M., Drews, A.,Stammen, S., Buchholz, K.,Dijkhuizen, L., and Dieter, J. "''A Bacillus megaterium Plasmid System for the Production, Export, and One-Step Purification of Affinity-Tagged Heterologous Levansucrase from Growth Medium". ''Appl Environ Microbiol.''. 2006. Volume 72. p. 1677–1679.] | |||
[2] [http://onlinelibrary.wiley.com/doi/10.1111/j.1365-2672.2005.02738.x/full Ding, Y., Wang, J., Liu, Y., and Chen,S. "''Isolation and identification of nitrogen-fixing bacilli from plant rhizospheres in Beijing region". ''Journal of Applied Microbiology''. 2005. Volume 99. p. 1271–1281.] | |||
[3] [http://jb.asm.org/content/193/16/4199.full.pdf+html Mark, E., et. al. "''Genome Sequences of the Biotechnologically Important Bacillus megaterium Strains QM B1551 and DSM319". ''Journal of Bacteriology''. 2011. Volume 193. p. 4199–4213.] | |||
[4] http://textbookofbacteriology.net/Bacillus_2.html | |||
[5] http://web.biosci.utexas.edu/psaxena/bio226r/pdf/3-43.pdf | |||
[6] Priest F. G., "Extracellular enzyme synthesis in the genus Bacillus." Bacteriological Reviews 1977, Volume 41. p. 711–753. | |||
[7] http://www.tgw1916.net/Bacillus/megaterium.html | |||
==Author== | ==Author== | ||
Latest revision as of 17:13, 4 December 2015
Classification
Kingdom- Bacteria; Phylum- Firmicutes; Class- Bacilli; Order- Bacillales; Family- Bacillaceae; Genus- Bacillus; Species- Bacillus Megaterium
Our Blast results were inconclusive. Bacillus Megaterium is the "best guess" narrowed down from our biochemical results and cellular and colonial characteristics in the lab**
Species
Genus species
Bacillus Megaterium
(Our second choice was Bacillus Cereus)
Habitat Information
The soil organism was isolated from a partially shaded creek at the San Marcos Golf Course in San Marcos, Tx on 09/02/15. The air temperature was 90F/32.2C, humidity 49%, pressure 30.04, solar radiation UV N/A. The grid coordinates from NRCS are 29 53.6' 97 55.4'. Spores are known to occur in soil, dust, water and plants.
Description and Significance
This organism is a gram positive, endospore forming, rod shaped bacteria. The colonial morphology is circular, smooth in the center, 'grainy' on the outside, undulate margins, umbonate elevation, and opaque with a golden appearance. The cellular arrangement is bacillus.
Kirby Bauer Antimicrobial results:
Strong Zone of Inhibition for (1) linezolid, (2) cefamandole, (3) azlocillin, (4) vancomycin, (5) sufisozazole.
Weak Zone of Inhibition for (6) oxacillin.
Disinfectant Sensitivity Results: Strong Zone of Inhibition for (1) clove, (2) 10% lysol. Weak Zone of Inhibition for (3) tea tree oil, (4) 100% bleach, and (5) lavender.
Anti-microbial activity was observed on an S. aureus test patch plate.
B. megaterium is a significant organism because it can be used industrially as a commercial strain and expression host [1]. B. megaterium has important implications as a host for penicillin aminidase and vitamin B12 [3]. A major advantage is that B. megaterium can secrete proteins directly into the growth medium [6]. Some strains are capable of nitrogen fixation [2].
Genome Structure
The size and content of the genome are undetermined due to inconclusive Blast results. PCR and DNA sequencing did not yield an adequate sequence for organism identification. Therefore, our genome information is based off of a 'best guess' of what organism might be based on biochemical and staining results. S Ribosomal sequence for our soil organism is not available.
The industrially significant B. megaterium strain QMB1551 has a 5.1 Mbp chromosome that carries approximately 5,300 genes. QMB1551 contains 7 natural plasmid (spBM100 to pBM700) ranging from 5.4 kb to over 164 kb. The chromosome is circular with an average G/C content of 38.2%. In 2011, Eppinger, et. al. completed the genomic sequence of QMB1551 [3].
Cell Structure, Metabolism and Life Cycle
The vegetative surface of B. megaterium consists of a proteinaceous surface layer (S-layer) and a cell wall made of peptidiglycan, containing meso-diaminopimelic acid (DAP). The function of the proteinaceous surface layer, made of crystalline glycoprotein subunits, is unknown. It may be involved in bacterial metal interactions [4]. Capsules may be present but were not observed for our soil organism. B. megaterium is mobile via a peritrichous flagella.
B. megaterium utilizes aerobic metabolism but has been known to survive anaerobic conditions. B. megaterium can produce metabolically inactive endospores to survive harsh conditions. The life cycle involves the following [5]:
1) Vegetative cell 2) Vegetative forms a septum (1/3 of the length of the cell) and pumps DNA into septum 3) Endospore develops and then surrounded by keratin-like coat 4) Vegetative cell dies; endospore release 5) Free mature endospore 6) Endospore becomes a vegetative cell
This is a link to endospore staining of B. megaterium:
http://www.microbelibrary.org/images/tu/bac_meg_endosp1.jpg
Physiology and Pathogenesis
B. megaterium is a common bacteria found in soil, does not produce endotoxins, and is non-pathogenic.
Biochemical characteristics:
NEGATIVE RESULTS INCLUDE: *Starch Hydrolysis, *Gelatin Hydrolysis, Methyl Red-Voges Proskauer, *Citrate test, SIM (negative for sulfur reduction, indole production, nonmotile organism), nitrate reduction, Urea hydrolysis, Eosin methylene blue (no coliforms, lactose fermenters, no growth: meaning possible gram +), Hektoen Enteric (presumptive ID for Gram +), MacConkey Agar (Gram + evidence), Decarboxylation Test, Phenylalanine deaminase, and *Catalase.
POSITIVE RESULTS INCLUDE: Casein hydrolysis, DNA hydrolysis, Lipid hydrolysis, Triple sugar iron, oxidase, blood agar (beta- complete lysis of RBC), mannitol salt agar (weak positive), phenylethal alcohol, and bile esculin.
Interpretation of Results: Our bacteria is a gram positive organism that possesses the enzymes casease, DNase, ligase, fermentable enzymes, and cytochrome C oxidase. This organism can completely lyse red blood cells, and can weakly ferment mannitol.
- *indicates inconsistent result with literature, likely due to "best guess" method.
References
http://www.eol.org/pages/974186/overview
[4] http://textbookofbacteriology.net/Bacillus_2.html
[5] http://web.biosci.utexas.edu/psaxena/bio226r/pdf/3-43.pdf
[6] Priest F. G., "Extracellular enzyme synthesis in the genus Bacillus." Bacteriological Reviews 1977, Volume 41. p. 711–753.
[7] http://www.tgw1916.net/Bacillus/megaterium.html
Author
Page authored by Hsiang-Yuan Sung and Kaleen Timon, student of Prof. Kristine Hollingsworth at Austin Community College.