User:S4188552: Difference between revisions
| (105 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
Aimee Davidson | Aimee Davidson 41885527 | ||
Bench E | Bench E | ||
23 Sept 2016 | |||
<ref>MICR3004</ref> | <ref>MICR3004</ref> | ||
| Line 7: | Line 7: | ||
===Higher order taxa=== | ===Higher order taxa=== | ||
Bacteria - Firmicutes - Negativicutes - Selenomonadales - Veillonellaceae - Veillonella<sup>[[#References|[3]]]</sup> | |||
===Species=== | ===Species=== | ||
'''Species name''': ''Veillonella parvula'' (subspecies ''parvula'') <sup>[[#References|[3]]]</sup> | |||
'''Type strain''': ATCC 10790 = CCUG 5123 = DSM 2008 = JCM 12972 = NCTC 11810 <sup>[[#References|[3]]]</sup> | |||
==Description and significance== | ==Description and significance== | ||
''Veillonella parvula'' is one of thirteen species of the ''Veillonella'' genus and one of only six of these species to be found within the human oral cavity <sup>[[#References|[4]]]</sup>. ''V.parvula'' was originally dubbed ''Staphylococcus parvulus'', by Veillon and Zuber in 1898, who first isolated and cultured the bacteria from an infected human appendix <sup>[[#References|[5]]]</sup>. It was subsequently further described and renamed ''Veillonella parvula'' in 1933 by Prevot [11]. ''V. parvula'' is a small gram-negative cocci, typically between 0.3-0.5 microns in diameter -which can be identitified by its characteristic pair grouping and distinctive red fluorescence under UV light and certain growth conditions <sup>[[#References|[6]]]</sup>. As mentioned previously it is commonly found within the human oral cavity, specifically on the tongue, buccal mucosa and in the saliva, however it can also be found within the human gastrointestinal, genitourinary and respiratory tracks<sup>[[#References|[7]]]</sup>, <sup>[[#References|[8]]]</sup>. Initially, ''V. parvula'' was thought to be non-pathogenic accounting for up to 98 % of cultivable ''Veillonella'' species in healthy human oral cavities <sup>[[#References|[7]]]</sup>. However, it is now known that while it is typically not directly pathogenic it regularly coaggregates with other pathogenic species, such as streptococcus and is a key early colonizer for the formation of oral biofilms <sup>[[#References|[4]]]</sup>. | |||
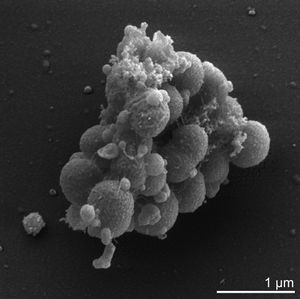
[[Image:Em_veillonella.jpeg|thumb|alt=em image.|Scanning electron micrograph image of ''V. parvula'' <sup>[[#References|[5]]]</sup>]] | |||
==Genome structure== | ==Genome structure== | ||
''V. parvula'' exhibits a circular 2.13 Mb (2,132,142 bp) genome, of which 88.46 % is DNA encoding, and was the first member of the ''Veillonella'' genus to have its complete genome sequenced. Its 1920 genes include: 1859 protein-encoding genes, 61 RNA specific genes and 15 pseudogenes, with an overall GC content of 38.63%. The majority of these genes are predicted to have either an unknown (385 genes) or general function (177 genes). Those genes with a predicted function are predominately involved with: translation, ribosomal structure, cell wall or membrane biogenesis, energy production and conversion, amino acid, coenzyme and inorganic transport and metabolism<sup>[[#References|[7]]]</sup>. | |||
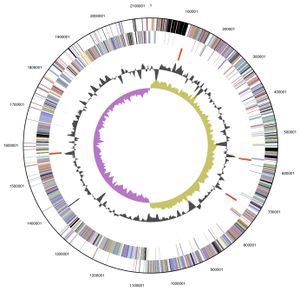
[[Image:Genome veillonella.jpeg|thumb|alt=genome image.|Genome structure of ''V. parvula'' <sup>[[#References|[7]]]</sup>.]] | |||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
Verkley ''et al'' established that 98 % of the phosphoglycerides that comprise ''V. parvula's'' cell membrane are either serine or ethanolamine based. These phosphoglycerides typically exist in single bonded carbon chains consisting of between 11 to 19 carbons, although chains containing 15,17,18 or 19 carbons atoms may also contain one double carbon bond [12]. Additionally, many of these phosphoglycerides exist in a plasmogen form which is thought to have an important role in the regulation of the cell's membrane fluidity<sup>[[#References|[6]]]</sup>,<sup>[[#References|[9]]]</sup>. The peptidoglycan cell wall of ''V. parvula'' consists of: glutamic acid in the D configuration; diaminopimelic acid in the meso configuration; and alpha-linked cadaverine and putrescine to glutamic acid<sup>[[#References|[7]]]</sup>. ''V. parvula'' lack flagella, spores and capsules and the ability to adhere to surfaces [6, 5]. It overcomes this lack of surface adhesion by adhering to specific surface structures on other bacterial species through lectin-carbohydrate mediated interactions<sup>[[#References|[7]]]</sup>. | |||
''V. parvula'' is unable to utilise carbohydrates, such as glucose, or amino acids in its energy production but rather utilises the metabolic end products of closely associated bacteria within the environment, particularly lactic acid producing species<sup>[[#References|[7]]]</sup>,<sup>[[#References|[5]]]</sup>. That is,it is able to metabolise lactate, pyruvate, malate and fumarate to produce propionate, acetic acid, carbon dioxide (CO2) and hydrogen<sup>[[#References|[7]]]</sup>. In addition, Ng and Hamilton established that ''V. parvula'' produces greater quantities of CO2 and propionate, which is explicitly shown in the following general equation of lactate metabolism: 8 lactate metabolises to 5 propionate + 3 acetate + 3 CO2 + H2 | |||
==Ecology== | ==Ecology== | ||
''V. parvula'' is a strict anaerobe that ideally grows at 37C and a pH range of 6.5 to 8.0, as such humans present as the ideal host. It is most commonly found within the human oral cavity and gastrointestinal tract, although it may also be found within the genitourinary and respiratory tracks<sup>[[#References|[7]]]</sup>,<sup>[[#References|[8]]]</sup>. Owing to ''V. parvula''’s inability to directly adhere to host surfaces it has no direct microbe to host interactions, rather it relies upon interactions with other microbes within the same biological niche<sup>[[#References|[5]]]</sup>. Specifically it is commonly a key early colonizer in early oral biofilm formation and facilitates the colonizing of middle and late phase pathogenic colonizers responsible for direct host/microbe interactions<sup>[[#References|[4]]]</sup>. | |||
==Pathology== | ==Pathology== | ||
''V. parvula'' has a well-established association with human oral diseases, in particular demonstrating a strong association with severe early childhood carries and intraradicular infections<sup>[[#References|[4]]]</sup>,<sup>[[#References|[7]]]</sup>. Recent research has also revealed that ''V. parvula'' is the predominant ''Veillonella'' species within dentine biofilms<sup>[[#References|[11]]]</sup>. Although ''V. parvula'' generally presents as a commensal or non-pathogenic resident in the bacterial communities of the oropharynx -, the gastrointestinal, genitourinary and respiratory tracks it has rarely been associated with a number of serious and life-threatening infections. These infections include: osteomyeltitis, spondylodiscitis, discitis, meningitis and sinus infections<sup>[[#References|[6]]]</sup>,<sup>[[#References|[8]]]</sup>. It has demonstrated resistance against a wide range of antibiotics including: tetracycline, erythromycin, gentamicin, karamycin, chloramphenicol and lincomycin. Although importantly at the time of its genome sequencing in 2010 it was still susceptible to penicillin, cephalotin and clindamycin<sup>[[#References|[7]]]</sup>. | |||
==Application to biotechnology== | ==Application to biotechnology== | ||
Perhaps largely owing to ''V. parvula''’s largely commensal nature, research thus far has not focused upon exploring its use in the field of bioengineering. Although a recent study by Periasamy and Kolenbrander has highlighted its potential as therapeutic target particularly with regards to dental plaque formation. They revealed that ''V. parvula'' was especially required for the development of three species biofilms. Thereby suggesting that prevention of ''V. parvula'' colonisation may hinder the development of these biofilms that contribute to human oral disease<sup>[[#References|[12]]]</sup>. | |||
==Current research== | ==Current research== | ||
A key advancement towards the study of ''V. parvula'' was made recently in June 2016. Historically species specific identification of members of ''Veillonella'' genus has been difficult owing to a lack of sufficiently reliable phenotypic and biochemical tests. Particularly the well-established identification method using 16S rDNA technology has proved unreliable. Mashima ''et al'' was able to overcome this issue by developing a novel one-step multiplex PCR method utilising purpose designed primer sets that target regions within the ''rpoB'' gene of individual ''Veillonella'' species<sup>[[#References|[4]]]</sup>. | |||
==References== | ==References== | ||
| Line 65: | Line 56: | ||
2. [http://www.homd.org Human Oral Microbiome] | 2. [http://www.homd.org Human Oral Microbiome] | ||
3. [http://www. | 3. [http://www.bacterio.net/-classificationsz.html#Veillonella List of prokaryotic names with standing in nomenclature] | ||
4. [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4915624/ Mashima, I., Theodorea, C.F., Thaweboon, B., Thaweboon, S., and Nakazawa, F. (2016) Identification of Veillonella Species in the Tongue Biofilm by Using a Novel One-Step Polymerase Chain Reaction Method. PLoS One. <b>11</b>(6):e0157516] | |||
5. [https://www-ncbi-nlm-nih-gov.ezproxy.library.uq.edu.au/pubmed/3904599 Delwiche, E.A., Pestka, J.J., Tortorello, M.L. (1985) The veillonellae: gram-negative cocci with a unique physiology. Ann. Rev. Microbiol <b>39</b>:175-93] | |||
6. [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC236067/ Verkley, A.J., Ververgaert, P.H., Prins, R.A., van Golde, L.M. (1975) Lipid-phase transitions of the strictly anaerobic bacteria Veillonella parvula and Anaerovibrio lipolytica. J Bacteriol <b>124</b>(3):1522-8] | |||
7. [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3035260/ Gronow, S., Welnitz, S., Lapidus, A., Nolan, M., Ivanova, N., Del Rio, T.G., Copeland, A., Chen, F., Tice, H., Pitluck, S., Cheng, J-F., Saunders, E., Brettin, T., Han, C., Detter, J.C., Bruce, D., Goodwin, L., Land, M., Hauser, L., Chang, Y-J., Jeffries, C.D., Pati, A., Mavromatis, K., Mikhailova, N., Chen, A., Palaniappan, K., Chain, P., Rohde, M., Göker, M., Bristow, J., Eisen, J.A., Markowitz, V., Hugenholtz, P., Kyrpides, N.C., Klenk, H-P., and Lucas. S. (2010) Complete genome sequence of Veillonella parvula type strain (Te3). Stand Genomic Sci <b>2</b>(1): 65-74.] | |||
8. [http://www.e-jmii.com/article/S1684-1182(14)00098-X/fulltext Chen, Y.C., Ko, P.H., Yang, C.J., Chen, Y.C., Tsai, C.C., Hsieh, M.H. (2014) Epidural abscess caused by Veillonella parvula: Case report and review of the literature. J Microbiol Immunol Infect. pii:S1684-1182(14)00098-X] | |||
9. [http://cid.oxfordjournals.org/content/31/3/839.long Bhatti, M.A., and Frank, M.O. (2000) Veillonella parvula meningitis: case report and review of Veillonella infections. Clin Infect Dis. <b>31</b>(3):839-40.] | |||
10. [http://www.ncbi.nlm.nih.gov/pubmed/4323300 Ng, S.K., and Hamilton, I.R. (1971) Lactate metabolism by Veillonella parvula. J Bacteriol. <b>105</b>(3):999-1005] | |||
11. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4374535/ Thuy, D., Sheehy E.C., Tonnie, M., Hughes, F., and Beighton, D. (2015) Transcriptomic analysis of three Veillonella spp. present in carious dentine and in the saliva of caries-free individuals. Front Cell Infect Microbiol. <b>5</b:25.] | |||
12. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2901697/ Periasamy, S., and Kolenbrander, P.E. (2010) Central role of the early colonizer Veillonella sp. in establishing multispecies biofilm communities with initial, middle, and late colonizers of enamel. J Bacteriol <b>192</b>(12):2965-2972] | |||
<references/> | <references/> | ||
This page is written by Aimee Davidson for the MICR3004 course, Semester 2, 2016 | This page is written by Aimee Davidson 41885527 for the MICR3004 course, Semester 2, 2016 | ||
Latest revision as of 12:38, 23 September 2016
Aimee Davidson 41885527 Bench E 23 Sept 2016 [1]
Classification
Higher order taxa
Bacteria - Firmicutes - Negativicutes - Selenomonadales - Veillonellaceae - Veillonella[3]
Species
Species name: Veillonella parvula (subspecies parvula) [3]
Type strain: ATCC 10790 = CCUG 5123 = DSM 2008 = JCM 12972 = NCTC 11810 [3]
Description and significance
Veillonella parvula is one of thirteen species of the Veillonella genus and one of only six of these species to be found within the human oral cavity [4]. V.parvula was originally dubbed Staphylococcus parvulus, by Veillon and Zuber in 1898, who first isolated and cultured the bacteria from an infected human appendix [5]. It was subsequently further described and renamed Veillonella parvula in 1933 by Prevot [11]. V. parvula is a small gram-negative cocci, typically between 0.3-0.5 microns in diameter -which can be identitified by its characteristic pair grouping and distinctive red fluorescence under UV light and certain growth conditions [6]. As mentioned previously it is commonly found within the human oral cavity, specifically on the tongue, buccal mucosa and in the saliva, however it can also be found within the human gastrointestinal, genitourinary and respiratory tracks[7], [8]. Initially, V. parvula was thought to be non-pathogenic accounting for up to 98 % of cultivable Veillonella species in healthy human oral cavities [7]. However, it is now known that while it is typically not directly pathogenic it regularly coaggregates with other pathogenic species, such as streptococcus and is a key early colonizer for the formation of oral biofilms [4].
Genome structure
V. parvula exhibits a circular 2.13 Mb (2,132,142 bp) genome, of which 88.46 % is DNA encoding, and was the first member of the Veillonella genus to have its complete genome sequenced. Its 1920 genes include: 1859 protein-encoding genes, 61 RNA specific genes and 15 pseudogenes, with an overall GC content of 38.63%. The majority of these genes are predicted to have either an unknown (385 genes) or general function (177 genes). Those genes with a predicted function are predominately involved with: translation, ribosomal structure, cell wall or membrane biogenesis, energy production and conversion, amino acid, coenzyme and inorganic transport and metabolism[7].
Cell structure and metabolism
Verkley et al established that 98 % of the phosphoglycerides that comprise V. parvula's cell membrane are either serine or ethanolamine based. These phosphoglycerides typically exist in single bonded carbon chains consisting of between 11 to 19 carbons, although chains containing 15,17,18 or 19 carbons atoms may also contain one double carbon bond [12]. Additionally, many of these phosphoglycerides exist in a plasmogen form which is thought to have an important role in the regulation of the cell's membrane fluidity[6],[9]. The peptidoglycan cell wall of V. parvula consists of: glutamic acid in the D configuration; diaminopimelic acid in the meso configuration; and alpha-linked cadaverine and putrescine to glutamic acid[7]. V. parvula lack flagella, spores and capsules and the ability to adhere to surfaces [6, 5]. It overcomes this lack of surface adhesion by adhering to specific surface structures on other bacterial species through lectin-carbohydrate mediated interactions[7]. V. parvula is unable to utilise carbohydrates, such as glucose, or amino acids in its energy production but rather utilises the metabolic end products of closely associated bacteria within the environment, particularly lactic acid producing species[7],[5]. That is,it is able to metabolise lactate, pyruvate, malate and fumarate to produce propionate, acetic acid, carbon dioxide (CO2) and hydrogen[7]. In addition, Ng and Hamilton established that V. parvula produces greater quantities of CO2 and propionate, which is explicitly shown in the following general equation of lactate metabolism: 8 lactate metabolises to 5 propionate + 3 acetate + 3 CO2 + H2
Ecology
V. parvula is a strict anaerobe that ideally grows at 37C and a pH range of 6.5 to 8.0, as such humans present as the ideal host. It is most commonly found within the human oral cavity and gastrointestinal tract, although it may also be found within the genitourinary and respiratory tracks[7],[8]. Owing to V. parvula’s inability to directly adhere to host surfaces it has no direct microbe to host interactions, rather it relies upon interactions with other microbes within the same biological niche[5]. Specifically it is commonly a key early colonizer in early oral biofilm formation and facilitates the colonizing of middle and late phase pathogenic colonizers responsible for direct host/microbe interactions[4].
Pathology
V. parvula has a well-established association with human oral diseases, in particular demonstrating a strong association with severe early childhood carries and intraradicular infections[4],[7]. Recent research has also revealed that V. parvula is the predominant Veillonella species within dentine biofilms[11]. Although V. parvula generally presents as a commensal or non-pathogenic resident in the bacterial communities of the oropharynx -, the gastrointestinal, genitourinary and respiratory tracks it has rarely been associated with a number of serious and life-threatening infections. These infections include: osteomyeltitis, spondylodiscitis, discitis, meningitis and sinus infections[6],[8]. It has demonstrated resistance against a wide range of antibiotics including: tetracycline, erythromycin, gentamicin, karamycin, chloramphenicol and lincomycin. Although importantly at the time of its genome sequencing in 2010 it was still susceptible to penicillin, cephalotin and clindamycin[7].
Application to biotechnology
Perhaps largely owing to V. parvula’s largely commensal nature, research thus far has not focused upon exploring its use in the field of bioengineering. Although a recent study by Periasamy and Kolenbrander has highlighted its potential as therapeutic target particularly with regards to dental plaque formation. They revealed that V. parvula was especially required for the development of three species biofilms. Thereby suggesting that prevention of V. parvula colonisation may hinder the development of these biofilms that contribute to human oral disease[12].
Current research
A key advancement towards the study of V. parvula was made recently in June 2016. Historically species specific identification of members of Veillonella genus has been difficult owing to a lack of sufficiently reliable phenotypic and biochemical tests. Particularly the well-established identification method using 16S rDNA technology has proved unreliable. Mashima et al was able to overcome this issue by developing a novel one-step multiplex PCR method utilising purpose designed primer sets that target regions within the rpoB gene of individual Veillonella species[4].
References
References examples
3. List of prokaryotic names with standing in nomenclature
- ↑ MICR3004
This page is written by Aimee Davidson 41885527 for the MICR3004 course, Semester 2, 2016
