User:S4263537: Difference between revisions
| (30 intermediate revisions by the same user not shown) | |||
| Line 26: | Line 26: | ||
The organism currently known as ''Veillonella parvula'' was discovered in 1898 by Veillon and Zuber who called it ''Staphylococcus parvulus''. However, it was in 1933 when Prevot split it off as the type member of the Veillonella genera1<sup>[[#References|[2]]]</sup>. . | The organism currently known as ''Veillonella parvula'' was discovered in 1898 by Veillon and Zuber who called it ''Staphylococcus parvulus''. However, it was in 1933 when Prevot split it off as the type member of the Veillonella genera1<sup>[[#References|[2]]]</sup>. . | ||
''Veillonella parvula'' are anaerobic gram negative cocci that are commonly found in oral cavities, the vagina and the gastrointestinal tract. Despite been a gram negative bacteria, as members of the Firmicutes Phylum, they’re actually more closely related to Gram Positive genera like ''Sporomusa'', ''Selenomonas'' or even ''Clostridium'', than they are to most gram negative genera<sup>[[#References|[ | ''Veillonella parvula'' are anaerobic gram negative cocci that are commonly found in oral cavities, the vagina and the gastrointestinal tract. Despite been a gram negative bacteria, as members of the Firmicutes Phylum, they’re actually more closely related to Gram Positive genera like ''Sporomusa'', ''Selenomonas'' or even ''Clostridium'', than they are to most gram negative genera<sup>[[#References|[2]]]</sup>. In particular, they share the presence of cadverine & putrescine in their cell walls with those close relatives like ''Sporomusa'' ans ''Selenomonas''. | ||
This organism glows red under a UV light<sup>[[#References|[ | This organism glows red under a UV light<sup>[[#References|[2]]]</sup> and has been successfully cultured on several types of media including trypticase soy agars (with sheep’s blood)<sup>[[#References|[3]]]</sup>, thioglycerate broth<sup>[[#References|[3]]]</sup>, and cystine-lactose-electrolyte deficient media<sup>[[#References|[4]]]</sup>. | ||
==Genome structure== | ==Genome structure== | ||
According to KEGG , the number of nucleotides in the V. parvula genome is around 2132142, the number of protein encoding genes is 1844 and there are also 64 RNA genes present.<sup>[[#References|[ | According to KEGG , the number of nucleotides in the V. parvula genome is around 2132142, the number of protein encoding genes is 1844 and there are also 64 RNA genes present.<sup>[[#References|[5]]]</sup> | ||
Known plasmids shared by Veillonella include: R plasmids for antibiotic-resistance genes<sup>[[#References|[ | Known plasmids shared by Veillonella include: R plasmids for antibiotic-resistance genes<sup>[[#References|[6]]]</sup>, and OK1 plasmid pVJL1 (the first shuttle vector designed for use on Veillonellae)<sup>[[#References|[7]]]</sup> | ||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
| Line 40: | Line 40: | ||
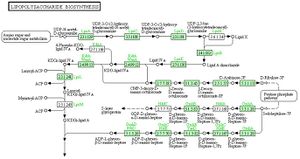
[[Image:V.parvula_LPS_Biosynthesis.jpeg|thumb|A KEGG Map that shows thee LPS Biosynthesis pathway as it exists in ''Veillonella parvula'' (with green boxes been genes present, white boxes been genes absent]]. | [[Image:V.parvula_LPS_Biosynthesis.jpeg|thumb|A KEGG Map that shows thee LPS Biosynthesis pathway as it exists in ''Veillonella parvula'' (with green boxes been genes present, white boxes been genes absent]]. | ||
''Veillonella parvula'' is a gram negative anaerobic, non-motile, non-sporulating mesophilic coccus<sup>[[#References|[ | ''Veillonella parvula'' is a gram negative anaerobic, non-motile, non-sporulating mesophilic coccus<sup>[[#References|[2]]]</sup>. It is known to be capable of reducing nitrate so as to produce arginine dihydrolase | ||
Whilst ''V.parvula'' is incapable of attaching to surfaces itself, it is able to attach to surface structures produced by other bacterial species and become a coloniser of those biofilms. This organism is an early-coloniser of subgingival biofilms (otherwise known as dental plaques) formed by members of the ''Streptococcus'' genus, and the ''Veillonella'' genera is not only a typical early coloniser of Streptococci-formed biofilms, but also tends to be co-aggregate with various Streptococci species in said biofilms<sup>[[#References|[ | Whilst ''V.parvula'' is incapable of attaching to surfaces itself, it is able to attach to surface structures produced by other bacterial species and become a coloniser of those biofilms. This organism is an early-coloniser of subgingival biofilms (otherwise known as dental plaques) formed by members of the ''Streptococcus'' genus, and the ''Veillonella'' genera is not only a typical early coloniser of Streptococci-formed biofilms, but also tends to be co-aggregate with various Streptococci species in said biofilms<sup>[[#References|[8]]]</sup>. | ||
''V. parvula'' is incapable of catabolising sugars and relies on the fermentation of organic acids such as lactate, succinate, malate and fumarate. As members of the ''Streptococci'' genera regularly produce these molecules as by-products of sugar catabolism, this is a potential explanation for why ''V.parvula'' is an early coloniser of Streptococci-formed biofilms. The by-products of the various fermentation reactions performed by ''V.parvula'' tend to include propionic acid, hydrogen, acetic acid and carbon dioxide. | ''V. parvula'' is incapable of catabolising sugars and relies on the fermentation of organic acids such as lactate, succinate, malate and fumarate. As members of the ''Streptococci'' genera regularly produce these molecules as by-products of sugar catabolism, this is a potential explanation for why ''V.parvula'' is an early coloniser of Streptococci-formed biofilms. The by-products of the various fermentation reactions performed by ''V.parvula'' tend to include propionic acid, hydrogen, acetic acid and carbon dioxide. | ||
| Line 50: | Line 50: | ||
==Ecology== | ==Ecology== | ||
''V. parvula'' is a moderate anaerobe<sup>[[#References|[ | ''V. parvula'' is a moderate anaerobe<sup>[[#References|[9]]]</sup> and while it prefers anaerobic environments in the human body, it has been shown to be capable of surviving for 72 hours under atmospheric oxygen conditions , though it can’t reproduce above an oxygen concentration of 2.5%<sup>[[#References|[9]]]</sup> (hence why it’s considered to be a moderate anaerobe, as strict anaerobes can’t survive oxygen concentrations above 0.5%). | ||
In the oral cavity, ''V. parvula'' will prefer subgingival biofilms. Other environments in which V.parvula can be found include upper respiratory tract, small intestines, and vagina<sup>[[#References|[ | In the oral cavity, ''V. parvula'' will prefer subgingival biofilms. Other environments in which ''V.parvula'' can be found include upper respiratory tract, small intestines, and vagina<sup>[[#References|[10]]]</sup>. Some studies indicate that ''V. parvula'' may be absent from tongue biofilms. | ||
''V. parvula'' tends to live in co-aggregations with other bacteria inside biofilms, including but not restricted to ''Streptococcus sanguinis'', ''Actinomyces naeslundii'', ''Actinomyces viscosus'', ''Streptococcus morbillorum''<sup>[[#References|[ | ''V. parvula'' tends to live in co-aggregations with other bacteria inside biofilms, including but not restricted to ''Streptococcus sanguinis'', ''Actinomyces naeslundii'', ''Actinomyces viscosus'', ''Streptococcus morbillorum''<sup>[[#References|[11]]]</sup>. and ''Actinomyces odontolyticus''<sup>[[#References|[12]]]</sup>. . | ||
The presence of ''V.parvula'' in biofilms formed by S. sanguinis is known to significantly increase the size of these biofilms in comparison to biofilms solely consisting of S.sanguinis . | The presence of ''V.parvula'' in biofilms formed by S. sanguinis is known to significantly increase the size of these biofilms in comparison to biofilms solely consisting of ''S.sanguinis''. | ||
There is some evidence that ''V. parvula'' might be unable to co-aggregate with ''Streptococcus salivarius''<sup>[[#References|[ | There is some evidence that ''V. parvula'' might be unable to co-aggregate with ''Streptococcus salivarius''<sup>[[#References|[11]]]</sup>., which could be due to the fact that ''S. salivarius'' is known to produce antimicrobial peptides known as BLIS (Bacteriocin-like Inhibitory Substances), which is known to inhibit ''Streptococcus pyogenes'' (the causative agent of strep throat). | ||
==Pathology== | ==Pathology== | ||
The most common disease consequence of ''V.parvula'' is in the formation of dental caries where they're often a component of larger microbial communities found by other bacteria such as members of ''Streptoccus'', and are associated with the 'purple complex' (along with Actinomyces odontolyticus) which is while not as associated with periodontitis as the red complex, is still part of the disease-causing biofilm<sup>[[#References|[11]]]</sup>. | |||
There is at least one case study of a Urinary Tract Infection (UTIs) caused by ''V. parvula'' which had symptoms including confusion, urinary retention, fever, general weakness and leucocytosis. This case study indicates that ''V. parvula'' UTIs can be treated with metronidazole, carbapenems and ceftriaxone<sup>[[#References|[4]]]</sup>. | |||
While rare, ''V. parvula'' has been implicated as the causative organisms in several meningitis cases, at least one of which was associated with severe sinusitis<sup>[[#References|[2]]]</sup>. Other organisms that might be co-infecting the host in cases of ''V. parvula''-associated meningitis include ''[[Prevotella intermedia]]'', ''[[Peptostreptococcus anaerobius]]'', ''[[Enterobacter cloacae]]'' and ''[[Propionibacterium acnes]]''. When ''V. parvula''-associated meningitis arises, treatment should include metronidazole and ceftriaxone. | |||
It has also been a known cause of chronic anaerobic pneumonitis when in a co-infection with ''[[Fusobacterium]]''<sup>[[#References|[12]]]</sup> . The symptomology of this disease include fever, haemoptysis and intermittent sneezing. The recommended antibiotics to use when ''Veillonella'' is a causative agent of pneumonitis include chloramphenicol, clindamycin, coamoxyclav, imipenem, and metronidazole. | |||
Other conditions that are known to be caused by ''V. parvula'' include osteomyelitis, bacteraemia (usually in association with osteomyelitis but not always<sup>[[#References|[13]]]</sup>), pelvic, lung and eye abcesses. | |||
Other conditions that are known to be caused by ''V. parvula'' include osteomyelitis, bacteraemia (usually in association with osteomyelitis but not always<sup>[[#References|[ | |||
==Application to biotechnology== | ==Application to biotechnology== | ||
| Line 77: | Line 75: | ||
The current literature up to 2016 doesn’t suggest that ''V. parvula'' is widely used for bioengineering or biotechnology applicatons. Therefore, this section will mostly focus on widely used drug targets for this organism. | The current literature up to 2016 doesn’t suggest that ''V. parvula'' is widely used for bioengineering or biotechnology applicatons. Therefore, this section will mostly focus on widely used drug targets for this organism. | ||
Metronidazole seems to be the most widely-used drug against ''V. parvula'' in the literature. It is of the 5-nitroimidazole class of antibiotics and is hypothesised to function by the following process: It first diffuses across cell membrane of the infectious agent. In anaerobes, the pyruvate-ferrodoxin oxidoreductase reduces it and alters its chemical structure in a way that promotes formation of cytotoxic free-radicals. These cytotoxic particles than interact with intracellular macromolecules such as the DNA and ultimately cause them to break<sup>[[#References|[ | Metronidazole seems to be the most widely-used drug against ''V. parvula'' in the literature. It is of the 5-nitroimidazole class of antibiotics and is hypothesised to function by the following process: It first diffuses across cell membrane of the infectious agent. In anaerobes, the pyruvate-ferrodoxin oxidoreductase reduces it and alters its chemical structure in a way that promotes formation of cytotoxic free-radicals. These cytotoxic particles than interact with intracellular macromolecules such as the DNA and ultimately cause them to break<sup>[[#References|[14]]]</sup>. These effects are rapid and don’t seem to be adversely impacted by other antibiotics nor does it adversely impact on other antibiotics, making it a very useful for multi-drug therapies. | ||
Due to ''V. parvula'' strains having acquired β-lactamase genes, it is only worth using β-lactam antibiotics if they are combined with something that will inhibit resistance in the organism, such as in coamoxyclav, where amoxicillin is combined with the β-lactamase inhibitor potassium clavulanate, which irreversibly binds the active site of β-lactamase, hence permanently inactivating it. | Due to ''V. parvula'' strains having acquired β-lactamase genes, it is only worth using β-lactam antibiotics if they are combined with something that will inhibit resistance in the organism, such as in coamoxyclav, where amoxicillin is combined with the β-lactamase inhibitor potassium clavulanate, which irreversibly binds the active site of β-lactamase, hence permanently inactivating it. | ||
| Line 84: | Line 82: | ||
Chloramphenicol & clindamycin both inhibit ribosomal peptidyl transferase activity in a way similar to macrolides, even though neither of them are macrolides. | Chloramphenicol & clindamycin both inhibit ribosomal peptidyl transferase activity in a way similar to macrolides, even though neither of them are macrolides. | ||
In some cases, tetracycline, which blocks translation by binding the 30S subunit of the ribosome. However, tetracycline resistance has been recorded since at least 1979<sup>[[#References|[ | In some cases, tetracycline, which blocks translation by binding the 30S subunit of the ribosome. However, tetracycline resistance has been recorded since at least 1979<sup>[[#References|[6]]]</sup>, and while some strains are still susceptible, resistance to tetracycline is wide-spread enough that other drugs are preferable. | ||
==Current research== | ==Current research== | ||
In 2015<sup>[[#References|[15]]]</sup>, a study on a mouse model of cystic fibrosis suggests that if both ''[[Pseuodomonas aeruginosa]]'' and ''V. parvula'' were co-infecting individuals with cystic fibrosis, then the presence of ''V. parvula'' will result in higher ''P. aeruginosa'' load in the hosts, resulting in worse clinical outcomes for those co-infected with both organisms. Within the cystic fibrosis tumours, ''V. parvula'' was found in the inner core of the tumour where it formed globular colonies whilst ''P. aeruginosa'' (an aerobic species) was found in the outer rim of the tumour tissue. However, as this was done in a mouse model, caution must be applied before applying these results to humans. | |||
In 2015, a study on a mouse model of cystic fibrosis suggests that if both ''[[Pseuodomonas aeruginosa]]'' and ''V. parvula'' were co-infecting individuals with cystic fibrosis, then the presence of ''V. parvula'' will result in higher ''P. aeruginosa'' load in the hosts, resulting in worse clinical outcomes for those co-infected with both organisms. Within the cystic fibrosis tumours, ''V. parvula'' was found in the inner core of the tumour where it formed globular colonies whilst ''P. aeruginosa'' (an aerobic species) was found in the outer rim of the tumour tissue. However, as this was done in a mouse model, caution must be applied before applying these results to humans. | |||
==References== | ==References== | ||
| Line 96: | Line 92: | ||
1. [http://www.bacterio.net/veillonella.html] | 1. [http://www.bacterio.net/veillonella.html] | ||
2. | 2. [Bhatti M.A., and Frank M.O., (2000) Veillonella parvula Meningitis: Case Report and Review of Veillonella Infections. ''Clinical Infectious Diseases: 31 839-840] | ||
3. [Gronow S, Welnitz S, Lapidus A, Nolan M, Ivanova N, et al. (2009) Complete genome sequence of Veillonella parvula type strain (Te3T) Standards in Genomic Sciences | 3. [Gronow S, Welnitz S, Lapidus A, Nolan M, Ivanova N, et al. (2009) Complete genome sequence of Veillonella parvula type strain (Te3T). ''Standards in Genomic Sciences'' 1:57-65 DOI:10.4056/sigs.521107] | ||
4. | 4. Berenger B.M., Chui L., Borkent A., and Lee M.C., (2015) Anaerobic urinary tract infection caused by Veillonella parvula identified using cystine-lactose-electrolyte deficient media and matrix-assisted laser desorption ionization-time of flight mass spectrometry. ID Cases 2 44-46 | ||
5. | 5. [http://www.genome.jp/kegg-bin/show_genomemap_top?org_id=vpr KEGG Genome map] | ||
6. | 6. Williams B.L., Osterberg S.K.A., and Jorgenson J 1979 Subgingival microfiora of periodontal patients on tetracycline therapy. ''Journal of Clinical Periodontology'': '''6'''. 210-221 | ||
7. [http://www.genome.jp/dbget-bin/www_bget?rs:NC_019271] | 7. [http://www.genome.jp/dbget-bin/www_bget?rs:NC_019271] | ||
8. | 8. Mashima I, Nakazawa F, The influence of oral Veillonella species on biofilms formed by Streptococcus species 2014 Anaerobe 28: 54-61 | ||
9.[Tally F.P., Stewart P.R., Sutter V.L., and Rosenblatt J.E., (1975) Oxygen Tolerance of Fresh Clinical Anaerobic Bacteria. Journal Clin Microbiol '''1(2)''': 161-164] | |||
10. [Hughes C.V., Kolenbrander P.E., Andersen R.N. and Moore L.V.H (1988) Coaggregation Properties of Human Oral Veilionella spp.: Relationship to Colonization Site and Oral Ecology. ''Am Soc. Mircobiol'' 1957-1963] | |||
11. Carrouell F., Viennot S., Santamaria J., Verber P. and Bourgeois D. (2016) Quantitative Molecular Detection of 19 Major Pathogens in the Interdental Biofilm of Periodontally Healthy Young Adults ''frontiers in Microbiol'' '''7'''(840) 1-16 doi: 10.3389/fmicb.2016.00840 | |||
12. A Shah, C Panjabi, V Nair, R Chaudhry, S.S. Thukral. (2008) Veillonella as a cause of chronic anaerobic pneumonitis. Journal of Infectious Diseases '''12''', 115-117 | |||
13. [Fisher RG, Denison MR (1996) Veillonella parvula Bacteremia without an Underlying Source. Am Soc. Microbiol 34-12. 3235-3236] | |||
14. [http://www.uptodate.com.ezproxy.library.uq.edu.au/contents/metronidazole-an-overview?source=search_result&search=metronidazole&selectedTitle=5%7E148#H2 Last accessed 22nd September, Last updated on the 31st May 2016, Author Melissa Johnson Website = UpToDate] | |||
15. Pustelny C., Komor U., Pawar V., Lorenz A., et al (2015) Contribution of ''Veillonella parvula'' to ''Pseudomonas aeruginosa'' - Mediated Pathogenicity in a Murine Tumor Model System. ''Infection and Immunity''. '''83(1)''': 417-429 | |||
<references/> | <references/> | ||
This page is written by <Kieran Maytom> for the MICR3004 course, Semester 2, 2016 | This page is written by <Kieran Maytom> for the MICR3004 course, Semester 2, 2016 | ||
Latest revision as of 21:56, 22 September 2016
Kieran Maytom Bench ID Date [1]
Classification
Higher order taxa
Domain = Bacteria
Phylum = Firmicutes
Class = Negativicutes
Order = Veillonellaes
Family = Veillonellaceae
Species
The species name is Veillonella parvula, with the type-strain been strain [ATCC 10790]
Description and significance
The organism currently known as Veillonella parvula was discovered in 1898 by Veillon and Zuber who called it Staphylococcus parvulus. However, it was in 1933 when Prevot split it off as the type member of the Veillonella genera1[2]. .
Veillonella parvula are anaerobic gram negative cocci that are commonly found in oral cavities, the vagina and the gastrointestinal tract. Despite been a gram negative bacteria, as members of the Firmicutes Phylum, they’re actually more closely related to Gram Positive genera like Sporomusa, Selenomonas or even Clostridium, than they are to most gram negative genera[2]. In particular, they share the presence of cadverine & putrescine in their cell walls with those close relatives like Sporomusa ans Selenomonas.
This organism glows red under a UV light[2] and has been successfully cultured on several types of media including trypticase soy agars (with sheep’s blood)[3], thioglycerate broth[3], and cystine-lactose-electrolyte deficient media[4].
Genome structure
According to KEGG , the number of nucleotides in the V. parvula genome is around 2132142, the number of protein encoding genes is 1844 and there are also 64 RNA genes present.[5]
Known plasmids shared by Veillonella include: R plasmids for antibiotic-resistance genes[6], and OK1 plasmid pVJL1 (the first shuttle vector designed for use on Veillonellae)[7]
Cell structure and metabolism
.
Veillonella parvula is a gram negative anaerobic, non-motile, non-sporulating mesophilic coccus[2]. It is known to be capable of reducing nitrate so as to produce arginine dihydrolase
Whilst V.parvula is incapable of attaching to surfaces itself, it is able to attach to surface structures produced by other bacterial species and become a coloniser of those biofilms. This organism is an early-coloniser of subgingival biofilms (otherwise known as dental plaques) formed by members of the Streptococcus genus, and the Veillonella genera is not only a typical early coloniser of Streptococci-formed biofilms, but also tends to be co-aggregate with various Streptococci species in said biofilms[8].
V. parvula is incapable of catabolising sugars and relies on the fermentation of organic acids such as lactate, succinate, malate and fumarate. As members of the Streptococci genera regularly produce these molecules as by-products of sugar catabolism, this is a potential explanation for why V.parvula is an early coloniser of Streptococci-formed biofilms. The by-products of the various fermentation reactions performed by V.parvula tend to include propionic acid, hydrogen, acetic acid and carbon dioxide.
One of the more important virulence factor is its Lipopolysaccharides, which are an integral part of the cell membrane and are responsible for much of the symptoms in bacteremia, meningitis & other rare disease implication of V. parvula.
Ecology
V. parvula is a moderate anaerobe[9] and while it prefers anaerobic environments in the human body, it has been shown to be capable of surviving for 72 hours under atmospheric oxygen conditions , though it can’t reproduce above an oxygen concentration of 2.5%[9] (hence why it’s considered to be a moderate anaerobe, as strict anaerobes can’t survive oxygen concentrations above 0.5%).
In the oral cavity, V. parvula will prefer subgingival biofilms. Other environments in which V.parvula can be found include upper respiratory tract, small intestines, and vagina[10]. Some studies indicate that V. parvula may be absent from tongue biofilms.
V. parvula tends to live in co-aggregations with other bacteria inside biofilms, including but not restricted to Streptococcus sanguinis, Actinomyces naeslundii, Actinomyces viscosus, Streptococcus morbillorum[11]. and Actinomyces odontolyticus[12]. .
The presence of V.parvula in biofilms formed by S. sanguinis is known to significantly increase the size of these biofilms in comparison to biofilms solely consisting of S.sanguinis. There is some evidence that V. parvula might be unable to co-aggregate with Streptococcus salivarius[11]., which could be due to the fact that S. salivarius is known to produce antimicrobial peptides known as BLIS (Bacteriocin-like Inhibitory Substances), which is known to inhibit Streptococcus pyogenes (the causative agent of strep throat).
Pathology
The most common disease consequence of V.parvula is in the formation of dental caries where they're often a component of larger microbial communities found by other bacteria such as members of Streptoccus, and are associated with the 'purple complex' (along with Actinomyces odontolyticus) which is while not as associated with periodontitis as the red complex, is still part of the disease-causing biofilm[11].
There is at least one case study of a Urinary Tract Infection (UTIs) caused by V. parvula which had symptoms including confusion, urinary retention, fever, general weakness and leucocytosis. This case study indicates that V. parvula UTIs can be treated with metronidazole, carbapenems and ceftriaxone[4].
While rare, V. parvula has been implicated as the causative organisms in several meningitis cases, at least one of which was associated with severe sinusitis[2]. Other organisms that might be co-infecting the host in cases of V. parvula-associated meningitis include Prevotella intermedia, Peptostreptococcus anaerobius, Enterobacter cloacae and Propionibacterium acnes. When V. parvula-associated meningitis arises, treatment should include metronidazole and ceftriaxone.
It has also been a known cause of chronic anaerobic pneumonitis when in a co-infection with Fusobacterium[12] . The symptomology of this disease include fever, haemoptysis and intermittent sneezing. The recommended antibiotics to use when Veillonella is a causative agent of pneumonitis include chloramphenicol, clindamycin, coamoxyclav, imipenem, and metronidazole.
Other conditions that are known to be caused by V. parvula include osteomyelitis, bacteraemia (usually in association with osteomyelitis but not always[13]), pelvic, lung and eye abcesses.
Application to biotechnology
The current literature up to 2016 doesn’t suggest that V. parvula is widely used for bioengineering or biotechnology applicatons. Therefore, this section will mostly focus on widely used drug targets for this organism.
Metronidazole seems to be the most widely-used drug against V. parvula in the literature. It is of the 5-nitroimidazole class of antibiotics and is hypothesised to function by the following process: It first diffuses across cell membrane of the infectious agent. In anaerobes, the pyruvate-ferrodoxin oxidoreductase reduces it and alters its chemical structure in a way that promotes formation of cytotoxic free-radicals. These cytotoxic particles than interact with intracellular macromolecules such as the DNA and ultimately cause them to break[14]. These effects are rapid and don’t seem to be adversely impacted by other antibiotics nor does it adversely impact on other antibiotics, making it a very useful for multi-drug therapies.
Due to V. parvula strains having acquired β-lactamase genes, it is only worth using β-lactam antibiotics if they are combined with something that will inhibit resistance in the organism, such as in coamoxyclav, where amoxicillin is combined with the β-lactamase inhibitor potassium clavulanate, which irreversibly binds the active site of β-lactamase, hence permanently inactivating it.
Cephalosporins such as ceftriaxone and carbapenems such as Imipenem are also used as an alternative to β-lactams due to targeting the same process (peptidoglycan synthesis) but are in general less susceptible to β-lactamases.
Chloramphenicol & clindamycin both inhibit ribosomal peptidyl transferase activity in a way similar to macrolides, even though neither of them are macrolides. In some cases, tetracycline, which blocks translation by binding the 30S subunit of the ribosome. However, tetracycline resistance has been recorded since at least 1979[6], and while some strains are still susceptible, resistance to tetracycline is wide-spread enough that other drugs are preferable.
Current research
In 2015[15], a study on a mouse model of cystic fibrosis suggests that if both Pseuodomonas aeruginosa and V. parvula were co-infecting individuals with cystic fibrosis, then the presence of V. parvula will result in higher P. aeruginosa load in the hosts, resulting in worse clinical outcomes for those co-infected with both organisms. Within the cystic fibrosis tumours, V. parvula was found in the inner core of the tumour where it formed globular colonies whilst P. aeruginosa (an aerobic species) was found in the outer rim of the tumour tissue. However, as this was done in a mouse model, caution must be applied before applying these results to humans.
References
1. [1]
2. [Bhatti M.A., and Frank M.O., (2000) Veillonella parvula Meningitis: Case Report and Review of Veillonella Infections. Clinical Infectious Diseases: 31 839-840]
3. [Gronow S, Welnitz S, Lapidus A, Nolan M, Ivanova N, et al. (2009) Complete genome sequence of Veillonella parvula type strain (Te3T). Standards in Genomic Sciences 1:57-65 DOI:10.4056/sigs.521107]
4. Berenger B.M., Chui L., Borkent A., and Lee M.C., (2015) Anaerobic urinary tract infection caused by Veillonella parvula identified using cystine-lactose-electrolyte deficient media and matrix-assisted laser desorption ionization-time of flight mass spectrometry. ID Cases 2 44-46
6. Williams B.L., Osterberg S.K.A., and Jorgenson J 1979 Subgingival microfiora of periodontal patients on tetracycline therapy. Journal of Clinical Periodontology: 6. 210-221
7. [2]
8. Mashima I, Nakazawa F, The influence of oral Veillonella species on biofilms formed by Streptococcus species 2014 Anaerobe 28: 54-61
9.[Tally F.P., Stewart P.R., Sutter V.L., and Rosenblatt J.E., (1975) Oxygen Tolerance of Fresh Clinical Anaerobic Bacteria. Journal Clin Microbiol 1(2): 161-164]
10. [Hughes C.V., Kolenbrander P.E., Andersen R.N. and Moore L.V.H (1988) Coaggregation Properties of Human Oral Veilionella spp.: Relationship to Colonization Site and Oral Ecology. Am Soc. Mircobiol 1957-1963]
11. Carrouell F., Viennot S., Santamaria J., Verber P. and Bourgeois D. (2016) Quantitative Molecular Detection of 19 Major Pathogens in the Interdental Biofilm of Periodontally Healthy Young Adults frontiers in Microbiol 7(840) 1-16 doi: 10.3389/fmicb.2016.00840
12. A Shah, C Panjabi, V Nair, R Chaudhry, S.S. Thukral. (2008) Veillonella as a cause of chronic anaerobic pneumonitis. Journal of Infectious Diseases 12, 115-117
13. [Fisher RG, Denison MR (1996) Veillonella parvula Bacteremia without an Underlying Source. Am Soc. Microbiol 34-12. 3235-3236]
15. Pustelny C., Komor U., Pawar V., Lorenz A., et al (2015) Contribution of Veillonella parvula to Pseudomonas aeruginosa - Mediated Pathogenicity in a Murine Tumor Model System. Infection and Immunity. 83(1): 417-429
- ↑ MICR3004
This page is written by <Kieran Maytom> for the MICR3004 course, Semester 2, 2016