Paracoccus denitrificans: Difference between revisions
No edit summary |
|||
| (23 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
{{Curated}} | |||
{{Biorealm Genus}} | {{Biorealm Genus}} | ||
| Line 5: | Line 6: | ||
===Higher order taxa=== | ===Higher order taxa=== | ||
Bacteria; Proteobacteria; Alpha Proteobacteria; Rhodobacterales; Rhodobacteraceae; | Eubacteria (Kingdom); Bacteria (Domain); Proteobacteria; Alpha Proteobacteria (Class); Rhodobacterales (Order); Rhodobacteraceae (Family); Paracoccus (Genus) | ||
===Species=== | ===Species=== | ||
''Paracoccus denitrificans'' | ''Paracoccus denitrificans'' | ||
| Line 18: | Line 14: | ||
==Description and significance== | ==Description and significance== | ||
Paracoccus denitrificans is a coccoid shaped gram negative bacteria. They live in the soil in either aerobic or anaerobic environments. They also have the ability to live in many different kinds of media including C1 and sulfur. They can either use organic energy sources, such as methanol or methylamine, or act as chemolithotrophs, using inorganic energy sources with carbon dioxide as their carbon source. Paracoccus denitrificans was first isolate in 1910 by Martinus Beijerinck, a Dutch microbiologist, and was given the name | ''Paracoccus denitrificans'' is a coccoid shaped gram-negative bacteria. They live in the soil in either aerobic or anaerobic environments. They also have the ability to live in many different kinds of media including C1 and sulfur. They can either use organic energy sources, such as methanol or methylamine, or act as chemolithotrophs, using inorganic energy sources with carbon dioxide as their carbon source. ''Paracoccus denitrificans'' was first isolate in 1910 by Martinus Beijerinck, a Dutch microbiologist, and was given the name ''Micrococcus denitrificans''. In 1969, D.H. Davis changed the name of the bacteria to its present name because of the discovery that the bacteria contained many features known to be in mitochondria. It is possible that ''Paracoccus denitrificans'' is an ancestor to the eukaryotic mitochondria. | ||
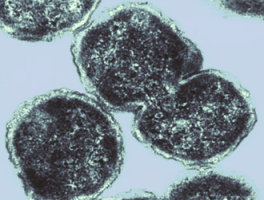
[[Image:P31.jpg]] | |||
Image taken by Richard Evans-Gowing at the University of East Anglia, Norwich, UK. | |||
==Genome structure== | ==Genome structure== | ||
The genome of Paracoccus denitrificans consists of two circular chromosomes and one plasmid. The first chromosome has 2,852,282 base pairs. The second chromosome has 1,730,097 base pairs and the plasmid has 653,815 base pairs. The plasmid encodes 611 known proteins such as Formyltetrahydrofolate deformylase and TonB-dependent siderophore receptor precursor. These proteins are not essential for the survival of the bacterium; however, the proteins transcribed and translated from the plasmid allow the bacterium to perform many of its metabolic functions. | The genome of ''Paracoccus denitrificans'' consists of two circular chromosomes and one plasmid. The first chromosome has 2,852,282 base pairs. The second chromosome has 1,730,097 base pairs and the plasmid has 653,815 base pairs. The plasmid encodes 611 known proteins such as Formyltetrahydrofolate deformylase and TonB-dependent siderophore receptor precursor. These proteins are not essential for the survival of the bacterium; however, the proteins transcribed and translated from the plasmid allow the bacterium to perform many of its metabolic functions. It is what gives ''Paracoccus denitrificans'' its unique features, such as the ability to metabolize ammonium to nitrogen gas. | ||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
The cell structures of Paracoccus denitrificans is similar to those in an eukaryotic mitochondria. They are able to obtain energy from both organic, such as methanol and methylamine, and inorganic compounds, such as hydrogen and sulfur. A | The cell structures of ''Paracoccus denitrificans'' is similar to those in an eukaryotic mitochondria. It is a gram-negative bacteria and therefore has all the properties typical of a gram-negative bacteria. This includes a double membrane with a cell wall. During the exponential growth phase, it is mainly rod-shaped; however, nearly spherical cells are observed when in the stationary phase. They are able to obtain energy from both organic, such as methanol and methylamine, and inorganic compounds, such as hydrogen and sulfur. A feature of this bacteria is its ability to single-handedly convert nitrate to dinitrogen in a process called denitrification. | ||
==Ecology== | ==Ecology== | ||
''Paracoccus denitrificans'' live primarily in the soil. They produce nitric oxide and nitrous oxide, which gives rise to atmospheric damage. They are also responsible for the loss of nitrogen fertilizer in agricultural soil. They do so by single-handedly converting nitrate into nitrogen gas. | |||
==Pathology== | ==Pathology== | ||
| Line 45: | Line 37: | ||
==Application to Biotechnology== | ==Application to Biotechnology== | ||
Paracoccus denitrificans produces | ''Paracoccus denitrificans'' produces more than 5000 proteins. Many of these proteins and enzymes are useful in biotechnological applications. One such a application is the construction of a bioreactor, in this case, a tubular gel containing two bacteria, for the removal of nitrogen from wastewater. ''Paracoccus denitrificans'' has the unusual ability of reducing nitrite to nitrogen gas. In this bioreactor, ''Paracoccus denitrificans'' is paired up with ''Nitrosomonas europaea'', which reduces ammonia to nitrite. This system simplifies the process of removing nitrogen from wastewater. | ||
==Current Research== | ==Current Research== | ||
A research study conducted in 2007 looked at the complex formed between cytochrome c oxidase. The lab used multi-frequency pulse electron paramagnetic resonance spectroscopy to study the complex. It was concluded that there was | A research study conducted in 2007 looked at the complex formed between cytochrome c and cytochrome c oxidase. The lab used multi-frequency pulse electron paramagnetic resonance spectroscopy to study the complex. It was concluded that there was no set orientation or distance between the two proteins that made up the complex. Another study, conducted in 2007, used Fourier transform infrared spectroscopy to study the effects of pH on the reduced-minus-oxidized FTIR spectra. This research study found two pH dependent processes. A third study on ''Paracoccus denitrificans'', published in 2007, studied the mechanism that reduces NO to N2O. The study concluded that the protons used in these reactions are taken up from the periplasm and not due to a proton electrochemical gradient. | ||
==References== | ==References== | ||
1. "Paracoccus denitrificans". 4 June 2007. <http://www.expasy.ch/sprot/hamap/PARDP.html> | |||
2. "CP000491" 4 June 2007 <http://www.expasy.org/cgi-bin/sprot-search-ful?makeWild=&SEARCH=CP000491> | |||
3. Reimann, J., Flock, U., Lepp, H., Honigmann, A., Adelroth, P. "A pathway for protons in nitric oxide reductase from Paracoccus denitrificans." Elsevier 1767.5 (2007), 362-373. 4 June 2007 <http://www.ncbi.nlm.nih.gov/sites/entrez?Db=pubmed&Cmd=ShowDetailView&TermToSearch=17466934> | |||
4. Gorbikova, E., Belevich, N., Wikström, M., Verkhovsky, M. "Protolytic reactions on reduction of cytochrome c oxidase studied by ATR-FTIR spectroscopy": Biochemistry. 46.13 (2007), 4177 - 4183. 4 June 2007 <http://www.ncbi.nlm.nih.gov/sites/entrez?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=17341097&query_hl=1&itool=pubmed_docsum> | |||
5. Lyubenova, S., Siddiqui, M., Vries, M., Ludwig, B., Prisner, T., "Protein-protein interactions studied by EPR relaxation measurements: cytochrome c and cytochrome c oxidase." J. Phys. Chem. B. 111.14 (2007), 3839 - 3846. 4 June 2007 <http://www.ncbi.nlm.nih.gov/sites/entrez?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=17388530&query_hl=1&itool=pubmed_docsum> | |||
6. "Paracoccus denitrificans". 4 June 2007. <http://www.jgi.doe.gov/> | |||
7. Uemoto, H., Saiki, H. "Nitrogen Removal by Tubular Gel Containing Nitrosomonas | |||
europaea and Paracoccus denitrificans." Applied and Environmental Microbiology. 62.11 (1996), 4224–4228. 4 June 2007 <http://aem.asm.org/cgi/reprint/62/11/4224?view=long&pmid=8900015> | |||
Edited by student of [mailto:ralarsen@ucsd.edu Rachel Larsen] and Kit Pogliano | Edited by student of [mailto:ralarsen@ucsd.edu Rachel Larsen] and Kit Pogliano | ||
KMG | |||
Latest revision as of 18:41, 22 April 2011
A Microbial Biorealm page on the genus Paracoccus denitrificans
Classification
Higher order taxa
Eubacteria (Kingdom); Bacteria (Domain); Proteobacteria; Alpha Proteobacteria (Class); Rhodobacterales (Order); Rhodobacteraceae (Family); Paracoccus (Genus)
Species
Paracoccus denitrificans
Description and significance
Paracoccus denitrificans is a coccoid shaped gram-negative bacteria. They live in the soil in either aerobic or anaerobic environments. They also have the ability to live in many different kinds of media including C1 and sulfur. They can either use organic energy sources, such as methanol or methylamine, or act as chemolithotrophs, using inorganic energy sources with carbon dioxide as their carbon source. Paracoccus denitrificans was first isolate in 1910 by Martinus Beijerinck, a Dutch microbiologist, and was given the name Micrococcus denitrificans. In 1969, D.H. Davis changed the name of the bacteria to its present name because of the discovery that the bacteria contained many features known to be in mitochondria. It is possible that Paracoccus denitrificans is an ancestor to the eukaryotic mitochondria.
Image taken by Richard Evans-Gowing at the University of East Anglia, Norwich, UK.
Genome structure
The genome of Paracoccus denitrificans consists of two circular chromosomes and one plasmid. The first chromosome has 2,852,282 base pairs. The second chromosome has 1,730,097 base pairs and the plasmid has 653,815 base pairs. The plasmid encodes 611 known proteins such as Formyltetrahydrofolate deformylase and TonB-dependent siderophore receptor precursor. These proteins are not essential for the survival of the bacterium; however, the proteins transcribed and translated from the plasmid allow the bacterium to perform many of its metabolic functions. It is what gives Paracoccus denitrificans its unique features, such as the ability to metabolize ammonium to nitrogen gas.
Cell structure and metabolism
The cell structures of Paracoccus denitrificans is similar to those in an eukaryotic mitochondria. It is a gram-negative bacteria and therefore has all the properties typical of a gram-negative bacteria. This includes a double membrane with a cell wall. During the exponential growth phase, it is mainly rod-shaped; however, nearly spherical cells are observed when in the stationary phase. They are able to obtain energy from both organic, such as methanol and methylamine, and inorganic compounds, such as hydrogen and sulfur. A feature of this bacteria is its ability to single-handedly convert nitrate to dinitrogen in a process called denitrification.
Ecology
Paracoccus denitrificans live primarily in the soil. They produce nitric oxide and nitrous oxide, which gives rise to atmospheric damage. They are also responsible for the loss of nitrogen fertilizer in agricultural soil. They do so by single-handedly converting nitrate into nitrogen gas.
Pathology
There are no known pathological effects of this bacterium on humans.
Application to Biotechnology
Paracoccus denitrificans produces more than 5000 proteins. Many of these proteins and enzymes are useful in biotechnological applications. One such a application is the construction of a bioreactor, in this case, a tubular gel containing two bacteria, for the removal of nitrogen from wastewater. Paracoccus denitrificans has the unusual ability of reducing nitrite to nitrogen gas. In this bioreactor, Paracoccus denitrificans is paired up with Nitrosomonas europaea, which reduces ammonia to nitrite. This system simplifies the process of removing nitrogen from wastewater.
Current Research
A research study conducted in 2007 looked at the complex formed between cytochrome c and cytochrome c oxidase. The lab used multi-frequency pulse electron paramagnetic resonance spectroscopy to study the complex. It was concluded that there was no set orientation or distance between the two proteins that made up the complex. Another study, conducted in 2007, used Fourier transform infrared spectroscopy to study the effects of pH on the reduced-minus-oxidized FTIR spectra. This research study found two pH dependent processes. A third study on Paracoccus denitrificans, published in 2007, studied the mechanism that reduces NO to N2O. The study concluded that the protons used in these reactions are taken up from the periplasm and not due to a proton electrochemical gradient.
References
1. "Paracoccus denitrificans". 4 June 2007. <http://www.expasy.ch/sprot/hamap/PARDP.html>
2. "CP000491" 4 June 2007 <http://www.expasy.org/cgi-bin/sprot-search-ful?makeWild=&SEARCH=CP000491>
3. Reimann, J., Flock, U., Lepp, H., Honigmann, A., Adelroth, P. "A pathway for protons in nitric oxide reductase from Paracoccus denitrificans." Elsevier 1767.5 (2007), 362-373. 4 June 2007 <http://www.ncbi.nlm.nih.gov/sites/entrez?Db=pubmed&Cmd=ShowDetailView&TermToSearch=17466934>
4. Gorbikova, E., Belevich, N., Wikström, M., Verkhovsky, M. "Protolytic reactions on reduction of cytochrome c oxidase studied by ATR-FTIR spectroscopy": Biochemistry. 46.13 (2007), 4177 - 4183. 4 June 2007 <http://www.ncbi.nlm.nih.gov/sites/entrez?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=17341097&query_hl=1&itool=pubmed_docsum>
5. Lyubenova, S., Siddiqui, M., Vries, M., Ludwig, B., Prisner, T., "Protein-protein interactions studied by EPR relaxation measurements: cytochrome c and cytochrome c oxidase." J. Phys. Chem. B. 111.14 (2007), 3839 - 3846. 4 June 2007 <http://www.ncbi.nlm.nih.gov/sites/entrez?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=17388530&query_hl=1&itool=pubmed_docsum>
6. "Paracoccus denitrificans". 4 June 2007. <http://www.jgi.doe.gov/>
7. Uemoto, H., Saiki, H. "Nitrogen Removal by Tubular Gel Containing Nitrosomonas europaea and Paracoccus denitrificans." Applied and Environmental Microbiology. 62.11 (1996), 4224–4228. 4 June 2007 <http://aem.asm.org/cgi/reprint/62/11/4224?view=long&pmid=8900015>
Edited by student of Rachel Larsen and Kit Pogliano
KMG