Aeromonas Hydrophila: Difference between revisions
No edit summary |
|||
| (52 intermediate revisions by 9 users not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | |||
{{Biorealm Genus}} | |||
==Classification== | |||
===Higher order taxa=== | |||
Cellular organism; Bacteria; Proteobacteria; Gammaproteobacteria; Aeromonadales; Aeromonadaceae; Aeromonas. | |||
===Species=== | |||
{| | |||
Some of the genes in Aeromonas hydrophila | | height="10" bgcolor="#FFDF95" | | ||
'''NCBI: [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Tree&id=2&lvl=3&lin=f&keep=1&srchmode=1&unlock Taxonomy]''' | |||
|} | |||
''Aeromonas hydrophila'' | |||
''Aeromonas hydrophila'' is a complex of three species including ''A.hydrophila sensu stricto'', ''A.bestiarum'', and ''A. salmonicida''. ''A.bestiarum'', and ''A. salmonicida'' are rarely present in humans [27]. | |||
Subspecies: ''Aeromonas hydrophila'' subsp. ''anaerogenes'', ''Aeromonas hydrophila'' subsp. ''decolorationis'', ''Aeromonas hydrophila'' subsp. ''dhakensis'', ''Aeromonas hydrophila'' subsp. ''hydrophila'' ATCC 7966, and ''Aeromonas hydrophila'' subsp. ''ranaei'' [19]. | |||
==Description and significance== | |||
[[image:Aeromonas2.jpg|frame|left|''Aeromonas hydrophila''. [http://www.nwfsc.noaa.gov/ Northwest Fisheries Science Center.] ]] | |||
''Aeromonas hydrophila'' is the most well known of the six species belonging to the genus ''Aeromonas''. It is rod- shaped, non- spore forming, oxidase- positive, glucose- fermenting, facultative anaerobic, gram- negative bacterium that inhabits aquatic environments [1, 2]. ''Aeromonas hydrophila'' colonies on agar plates are smooth, convex, and rounded, and they are tan/buff-colored on trypticase soy agar [25, 29]. This bacterium can be found in fresh, brackish, estuarine, marine, chlorinated and unchlorinated water supplies worldwide, with highest numbers obtained in the warmer climates [1, 2, 3, 4]. Though most commonly found in aquatic environments, ''Aeromonas hydrophila'' can also inhabit soil [25, 28]. The bacterium has optimal growth at 28°C but can also grow at the limits from 4°C to 37°C [1]. Some strains of ''Aeromonas hydrophila'' are capable of causing disease in fish and amphibians as well as in humans who may acquire infections through open wounds or by ingestion of an adequate number of the organisms in water or food. ''Aeromonas hydrophila'' has been isolated from raw vegetables like carrots, lettuce, and parsley. No outbreaks of this ''A. hydrophila'' have been associated with raw vegetables yet, but it does have the ability to grow on them even at cold, refrigeration temperature [26]. This bacterium can digest materials such as gelatin, hemoglobin, and elastin. ''Aeromonas hydrophila'' was isolated from diseased cold- and warm- blooded animals for over 100 years and from humans since the early 1950s [1, 5]. It is also hard to kill because it is a resistant bacterium. ''Aeromonas hydrophila'' is resistant to chlorine and refrigeration or cold temperatures [24]. | |||
==Genome structure== | |||
The complete genome of ''Aeromonas hydrophila'' strain ATCC 7966T was sequenced [14]. The genome is comprised of a single circular 4,744,448 bp chromosome with 61.5% GC content [20]. Its entire genome consists of 4,122 protein coding genes and 159 RNA genes: 128 tRNA genes amd 30 rRNA genes. It was possible to assign putative functions to 72.3% of the CDSs, while 21.5% possessed similarity to genes of unknown function, and no function could be proposed for 6.2% of the CDSs [21]. The complete genome sequence of ''Aeromonas hydrophila'' ATCC 7966T reveals mechanisms contributing to virulence and metabolic condition that allow the organism to grow in a variety of environments and explains how ''Aeromonas hydrophila'' is able to survive in polluted or oxygen- poor environments and to colonize and cause illness in humans and other hosts. However, two recognized virulence markers, a type III secretion system and a lateral flagellum, that are reported in other ''Aeromonas hydrophila'' strains are not identified in the sequenced isolate, ATCC 7966T. Given the ubiquity and free- living lifestyle of this organism, there is relatively little evidence of fluidity in terms of mobile elements in the genome of this particular strain. Thus, the ''Aeromonas hydrophila'' genome sequence provides valuable insights into its ability to flourish in both aquatic and host environments [14]. | |||
A number of virulence factors produced by ''Aeromonas'' species, however, their association with diarrhea have not been clearly linked [14]. Recently, a cytotoxic enterotoxin (Act), a heat- unstable cytotonic enterotoxin (Alt), and a heat- stable cytotonic enterotoxin (Ast) has been characterized from a diarrheal isolated of ''Aeromonas hydrophila''. The Act is a single- chain polypeptide that is one of the primary genes to make this species pathogenic. Act is aerolysin related and has hemolytic, cytotoxic, and enterotoxic activities. Alt and Ast also have genes that contribute to its toxicity, but are not related to cholera toxin [6]. | |||
==Cell structure and metabolism== | |||
''Aeromonas hydrophila'' are gram- negative straight rods with rounded ends. They usually grow from 0.3 to 1.0 μm in diameter and 1.0 to 3.5 μm in length. ''Aeromonas hydrophila'' does not form endospores, and can grow in temperatures as low as 4°C. This bacterium is motile by polar flagella. ''Aeromonas hydrophila'' are heterotrophic organisms. They can exist in both aerobic and anaerobic environments, hence it is a facultative aerobe that ferments both glucose and sucrose( both A/-). It is oxidase positive. ''Aeromonas hydrophila'' can also digest gelatin, hemoglobin, and elastin [23, 24]. | |||
''Aeromonas hydrophila'' has comprehensive biosynthetic abilities. The TCA cycle is complete and complemented by TCA cycle intermediates which can be replenished from acetate for use in various biosynthetic reactions. The Entner- Doudoroff and glycolytic pathways are intact, while the pentose phosphate pathway appears to be missing the oxidative branch. Complete multi- step pathways for synthesizing all amino acids are predicted, as are biosynthetic pathways for numerous cofactors such as biotin, glutathione, ubiquinone and menaquinone, pantothenate, thiamine, riboflavin, heme, molybdopterin, iron- sulfur clusters, coenzyme A, and tetrahydrofolate [14]. ''Aeromonas hydrophila'' has the oxygen- sensitive IscSUA- HscBA- Fdx system for the biosynthesis of iron- sulfur clusters rather than the oxygen- resistant SUF system, typically associated with aerobic and facultatively anaerobic organisms [14]. Under anaerobic growth conditions of ''Aeromonas hydrophila'', a NiFe uptake hydrogenase may act as electron carrier to appropriate acceptors, and immediately downstream is a putative nickel transporter, followed by various hydrogenase maturations. Additionally, nitrate is converted to nitrite and then to ammonia in the general assimilatory pathway. Nitrate may also act as an electron acceptor for anaerobic respiration [14]. | |||
Sulfate assimilation is accomplished by reduction of sulfate to sulfide with the participation of a sulfate adenylyltransferase, adenylylsulfate kinase, and phosphoadenosine phosphosulfate reductase. The sulfite product from the assimilation pathway is reduced by sulfite reductase to sulfide for incorporation into an amino acid, peptide, protein, etc. Additionally, ''Aeromonas hydrophila'' has a dissimilatory anaerobic sulfite reductase involved in dissimilation of oxidized anions for energy transduction. This enzyme catalyzes hydrogen sulfide production from sulfite, which is regulated by electron acceptors from hydrogen or an organic substrate, and serves as an important energy- conserving step. It also confers the ability to synthesize cysteine anaerobically. Other electron acceptor sources utilized by ''Aeromonas hydrophila'' include tetrathionate, fumarate, and trimethyl- N- oxide [14]. | |||
==Ecology== | |||
''Aeromonas hydrophila'' are ubiquitous bacteria which are found in a variety of aquatic environments worldwide, including bottled water, chlorinated water, well water, and heavily polluted waters. The organism is posted in the Contaminant Candidate List by the Environmental Protection Agency, and U.S. water supplies are routinely examined for it. Even ''Aeromonas hydrophila'' confer the metabolic versatility to persist in its aquatic habitats or that facilitate ecological interactions with other prokaryotic and eukaryotic organisms [14]. ''Aeromonas hydrophila'' infections elevate occurrence with environmental changes, in contaminated environments, change in the temperature, and when an organism is already infected with a virus or another bacterium [23]. | |||
==Pathology== | |||
''Aeromonas hydrophila'' causes a variety of diseases in both fish and human populations. The ubiquitous nature of the bacterium in aquatic environments provides significant opportunity for animals, mainly fish and amphibians to contact and ingest organisms [14]. | |||
''Aeromonas hydrophila'' is very toxic to many organisms because of its structure. When it enters the body of fish, amphibians, or humans, it travels via the bloodstream to the first available organ. It produces aerolysin cytotoxic enterotoxin (Act) which is one of the major virulence factors. Its toxin is produced and secreted by the cell from a type II secretion system. The toxin binds to high- affinity receptors and undergoes oligomerization to form a heptameric pore- forming complex which allows passage of small molecules in the plasma membrane, resulting in permeabilization of the cell, cell death, and eventually tissue destruction. ''Aeromonas hydrophila'' is also known as an opportunistic pathogenic bacterium, meaning they only infect hosts with weakened immune responses [6, 7]. Other virulence functions include a surface layer which inhibits complement- mediated killing, type IV pili for attachment, a set of extracellular proteases which can cause tissue damage, the ability to form capsules, and polar and lateral flagella [14, 25]. The polar flagella work to adhere this bacteria to surfaces, and the lateral flagella allow for colonization [25]. ''Aeromonas hydrophila'' also contains lipopolysaccharide on its outer membrane, which is a virulence factor in human infections [25]. Though ''Aeromonas hydrophila'' is considered a pathogenic bacterium, scientists have not been able to prove that it is the actual cause of some of the diseases it is associated with. It is believed that this bacterium aids in the infection of diseases, but do not cause the diseases themselves [14]. | |||
'''The disease in fish and amphibians''' | |||
''Aeromonas hydrophila'' cause illness mainly in fish and amphibians because this bacterium lives in aquatic environments. It is related to a disease found in frogs called red leg that causes internal or fatal hemorrhage. When infected with ''Aeromonas hydrophila'', fish develop ulcers, fin rot, tail rot, and hemorrhagic septicaemia [12, 13]. Specially, hemorrhagic septicaemia causes lesions that lead to scale shedding, hemorrhages in the gills and anal area, ulcers, exophthalmia, and abdominal swelling [1, 23]. A study of retail fish in Malaysia conducted in 2003 had a total of 87 market fish samples. 11.5% of these samples contained ''Aeromonas hydrophila''. They then looked at the antibiotic resistance patterns of the ''Aeromonas'' species in these fish and found that there was frequent resistance to ampicillin, carbenicillin, erythromycin, and streptomycin [28]. | |||
'''The disease in humans''' | |||
''Aeromonas hydrophila'' is also pathogenic to humans. It causes gastroenteritis which can affect anyone, but it most occurs in young children and people who have compromised immune systems or growth problems [12, 13]. This bacterium is linked to two types of gastroenteritis. The first type is a disease similar to cholera which causes rice- water diarrhea. Mild symptoms include fever and chills, but patients who become overwhelming bacterial infection with ''Aeromonas hydrophila'' often exhibit abdominal pain, nausea, vomiting, and diarrhea [1, 23]. | |||
The other type of disease is dysenteric gastroenteritis that causes loose stools filled with blood and mucus. Dysenteric gastroenteritis is the most severe out of the two types, and can last for multiple weeks. ''Aeromonas hydrophila'' is also associated with cellulitis, an infection that causes inflammation in the skin tissue. It also causes diseases such as myonecrosis and eczema in people with compromised immune systems [1, 14, 23]. | |||
'''Treatment''' | |||
The most common treatment for ''Aeromonas hydrophila'' infection in humans are broad-spectrum antibiotics, like tetracycline [25]. It is particularly susceptible to fluoroquinolones (a family of antibiotics). The most effective were levofloxacin, gatifloxacin, ciprofloxacin, and moxifloxacin. Resistance to these antibiotics is rare [27]. | |||
==Application to Biotechnology== | |||
''Aeromonas hydrophila'' is a gram- negative opportunistic pathogen in fish, amphibians, and humans. Many bacterial pathogens of animals and plants have been shown to inject anti- host virulence determinants into the hosts by a type III secretion system (TTSS). Degenerate primers based on ''lcrD'' family genes are present in every known TTSS that are allowed us to locate the TTSS gene cluster in ''Aeromonas hydrophila'' AH- 1 [15]. A series of genome walking steps facilitate to identify 25 open reading frames that encode proteins homologous to TTSSs in other bacteria. With the PCR- based analysis, it shows that the presence of ''lcrD'' homologs in all of the 33 strains of ''Aeromonas hydrophila'' isolated from various sources. Insertional inactivation of two of the TTSS genes led to decreased cytotoxicity in carp epithelial cells, increased phagocytosis, and reduced virulence in blue gourami. These results indicate that a TTSS is required for ''Aeromonas hydrophila'' pathogenesis. The TTSS identified may facilitate to develop suitable vaccines as well as to understand more of the pathogenesis of ''Aeromonas hydrophila'' [15]. | |||
Aeromonas hydrophila | |||
==Current Research== | |||
'''Aeromonas hydrophila AH- 3 AexT is an ADP-ribosylating toxin secreted through the type III secretion system [16].''' | |||
We cloned and sequenced an ADP- ribosylating toxin (AexT) from a mesophilic ''Aeromonas hydrophila'' strain AH-3 with a type III secretion system (T3SS). This toxin only showed homology, in genes and proteins, with the first half of ''Aeromonas salmonicida'' AexT. The ''Aeromonas hydrophila'' AexT showed ADP- ribosyltransferase activity, translocation through the T3SS system, and this ''Aeromonas hydrophila'' T3SS system is inducible under calcium- depleted conditions. The ''Aeromonas hydrophila'' aexT mutant showed a slight reduction in their virulence assayed by several methods when compared to the wild- type strain, while an ''Aeromonas hydrophila'' T3SS mutant is highly reduced in virulence on the same assays. The ''Aeromonas hydrophila'' AexT is the first described and the smallest T3SS effector toxin found in mesophilic ''Aeromonas'' with a functional T3SS [16]. | |||
Aeromonas hydrophila | |||
'''Further characterization of a type III secretion system (T3SS) and of a new effector protein from a clinical isolate of Aeromonas hydrophila- Part I [17].''' | |||
A type III secretion system (T3SS)- associated cytotoxin, AexT, with ADP-ribosyltransferase activity and homology to ''Pseudomonas aeruginosa'' bifuncational toxins ExoT/S, was recently identified from a fish pathogen ''Aeromonas salmonicida''. In this study, we reported the molecular characterization of an aexT- like toxin gene (designated as aexU) from a diarrheal isolate SSU of ''Aeromonas hydrophila''. The aexU gene was 1539 bp in length and encoded a protein of 512 amino acid residues. The NH(2)- terminus of AexU (aa residues 1-231) exhibited a 67% homology with the NH(2)-terminus of AexT from Aeromonas salmonicida. Importantly, its COOH- terminus (aa residues 232-512) had no homology with any known functional proteins in the database; however, the full-length AexU retained ADP- ribosyltransferase activity. The expression and subsequent secretion of AexU was T3SS dependent, as inactivation of the ascV gene that codes for an inner- membrane component of the T3SS channel from the wild- type bacterium, blocked translocation of AexU in HT-29 human colonic epithelial cells. We provided evidence that inactivation of acrV and axsE genes from ''Aeromonas hydrophila'' SSU, altered expression and/ or secretion of AexU. We deleted an aexU gene from the WT, as well as from the DeltaaopB mutant, of ''Aeromonas hydrophila'', generating a single knockout (DeltaaexU) and a double knockout mutant, DeltaaopB/ DeltaaexU. Increased phagocytosis was observed in RAW264.7 murine macrophages infected with the DeltaaopB/ DeltaaexU mutant, as compared to macrophages when infected with the parental DeltaaopB strain. Further, mice infected with the DeltaaexU mutant had a 60% survival rate, compared to animals infected with the WT or the DeltaaexU- complemented strain that caused 90- 100% of the animals to die at a 2-3 LD(50s) dose. Immunization of mice with the recombinant AexU protected them from subsequent lethal challenge dose by the WT bacterium. Finally, we detected specific anti- AexU antibodies in the sera of mice that survived challenge by the WT bacterium, which may indicate that AexU plays an important role in the pathogenesis of ''Aeromonas'' infections [17]. | |||
'''Biological characterization of a new type III secretion system effector from a clinical isolate of Aeromonas hydrophila- Part II [18].''' | |||
We recently identified a novel type III secretion system (T3SS) effector, AexU, from a diarrheal isolate SSU of ''Aeromonas hydrophila'', and demonstrated that mice infected with the DeltaaexU mutant were significantly protected from mortality. Although the NH(2)- terminal domain of this toxin exhibits homology to AexT of ''Aeromonas salmonicida'', a fish pathogen, and ExoT/ S of ''Pseudomonas aeruginosa'', the COOH-terminal domain of AexU is unique, with no homology to any known proteins in the NCBI database. In this study, we purified the full- length AexU and its NH(2)- terminal (amino acid residues 1-231) and COOH-terminal (amino acid residues 232-512) domains after expression of their corresponding genes in ''E.coli'' as histidine- tag fusion proteins using the bacteriophage T7 RNA polymerase/ promoter-based pET-30a vector system. The full- length and NH(2)- and COOH-terminal domains of AexU exhibited ADP-ribosyltransferase activity, with the former two exhibiting much higher activity than the latter. These different forms of AexU were also successfully expressed and produced in the HeLa Tet- Off cell system using a pBI-EGFP vector, as demonstrated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, Western blot analysis, and intracellular staining of the toxin using flow cytometric analysis. Production of AexU in HeLa cells resulted in possible actin reorganization and cell rounding, as determined by phalloidin staining and confocal microscopy. Based on electron microscopy, the toxin also caused chromatin condensation, which is indicative of apoptosis. Apoptosis of HeLa cells expressing and producing AexU was confirmed by 7- amino actinomycin D (7-AAD) and MTT [3- (4,5- dimethylthiazol-2- yl)- 2,5- diphenyl tetrasodium bromide] assays, by detection of cytoplasmic histone-associated DNA fragments, and by activation of caspases 3 and 9. These effects were much more pronounced in host cells that expressed and produced the full-length or NH(2)- terminal domain of AexU, compared to those that expressed and produced the COOH- terminal domain or the vector alone. This study represents the first characterization of this novel T3SS effector [18]. | |||
==References== | |||
1. Hayes, John. "Aeromonas hydrophila." Oregon State University. <http://hmsc.oregonstate.edu/classes/MB492/hydrophilahayes/> | |||
2. Hazen, T.C., Flierman, C.B., Hirsch, R.P. and Esch, G.W, “Prevalence and distribution of Aeromonas hydrophila in the United States” Appl. Environ. Microbiol, 1978, 36:731–738. | |||
3. Van der Kooj, D., “Properties of Aeromonads and their occurence and hygenic significance in drinking water”, Zentralb. Bakt. Hyg. B, 1988, 187:1–17. | |||
4. Kaper, J.B., Lockman, H., Colwell, R.R. and Joseph, S.W., “Aeromonas hydrophila: ecology and toxigenicity of isolates form an estuary” J. Appl. Microbiol, 1980, 50:359–377. | |||
5. Mathewson, J.J. and Dupont, H.L., “Aeromonas species: role as human pathogens”, In: Remington, J.S., Swartz, M.N. (eds.), “Current Clinical Topics in Infectious Diseases”, 1992, Vol12e. Cambridge: Blackwell Scientific. pp 26–36. | |||
6. Albert, M. John et. al. "Prevalence of Enterotoxin Genes in Aeromonas spp. Isolated from Children with Diarrhea, Healthy Controls, and the Environment." Journal of Clinical Microbiology, October 2000, p. 3785-3790, Vol. 38, No. 10. < http://jcm.asm.org/cgi/content/full/38/10/3785> | |||
7. Chopra, A.K., X.-J. Xu, D. Ribardo, M. Gonzalez, K. Kuhl, J. W. Peterson, and C. W. Houston. "The Cytotoxic Enterotoxin of Aeromonas hydrophila Induces Proinflammatory Cytokine Production and Activates Arachidonic Acid Metabolism in Macrophages." Infection and Immunity, May 2000, p. 2808-2818, Vol. 68, No.5. < http://iai.asm.org/cgi/content/full/68/5/2808> | |||
8. Cipriano, Rocco L. "Aeromonas hydrophila and Motile Aereomonad Septicemias of Fish." Fish and Disease Leaflet 68. 2001. < http://www.lsc.usgs.gov/FHB/leaflets/FHB68.pdf > | |||
9. Seetha KS, Jose BT, Jasthi A. "Meningitis due to aeromonas hydrophila." Indian J Med Microbiol 2004;22:191-192 < http://www.ijmm.org/article.asp?issn=0255-0857;year=2004;volume=22;issue=3;spage=191;epage=192;aulast=Seetha > | |||
10. Xu, Xin-J., M. R. Ferguson, V. L. Popov, C. W. Houston, J. W. Peterson, and A. K. Chopra. "Role of a Cytotoxic Enterotoxin in Aeromonas-Mediated Infections: Development of Transposon and Isogenic Mutants." Infect Immun, August 1998, p. 3501-3509, Vol. 66, No. 8. < http://iai.asm.org/cgi/content/full/66/8/3501 > | |||
11. Fulton, MacDonald. "THE BACTERIUM AEROMONAS HYDROPHILA FROM LIZARDS OF THE GENUS ANOLIS IN PUERTO RICO". Lousiana State University Medical Center, New Orleans, LA. <www.uprm.edu/publications/cjs/VOL05/P099-102.PDF>. | |||
12. "Aeromonas hydrophila." Bad Bug Book Foodborne Pathogenic Microorganisms and Natural Toxins Handbook. <http://vm.cfsan.fda.gov/~mow/chap17.html> | |||
13. "Aeromonas hydrophila and Related Bacteria." International Specialty Supply. <http://www.sproutnet.com/Reports/aeromonas_hydrophila.htm> | |||
14. Rekha Seshadri, Sam W. Joseph, Ashok K. Chopra, Jian Sha, Jonathan Shaw, Joerg Graf, Daniel Haft, Martin Wu, Qinghu Ren, M. J. Rosovitz, Ramana Madupu, Luke Tallon, Mary Kim, Shaohua Jin, Hue Vuong, O. Colin Stine, Afsar Ali, Amy J. Horneman, and John F. Heidelberg “Genome Sequence of Aeromonas hydrophila ATCC 7966T: Jack of All Trades”, J Bacteriol. 2006 December. <http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1698176> | |||
15. H. B. Yu, P. S. Srinivasa Rao, H. C. Lee, S. Vilches, S. Merino, J. M. Tomas, and K. Y. Leung, “A Type III Secretion System Is Required for Aeromonas hydrophila AH-1 Pathogenesis”, Infect Immun. 2004 March; 72(3): 1248–1256. | |||
<http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=356039> | |||
16. Vilches S, Wilhelms M, Yu HB, Leung KY, Tomas JM, Merino S “Aeromonas hydrophila AH-3 AexT is an ADP-ribosylating toxin secreted through the type III secretion system”, Microb Pathog. 2007 Jul 1; PMID: 17689917 <http://www.ncbi.nlm.nih.gov/sites/entrez?Db=pubmed&Cmd=ShowDetailView&TermToSearch=17689917&ordinalpos=3&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum> | |||
17. Sha J, Wang SF, Suarez G, Sierra JC, Fadl AA, Erova TE, Foltz SM, Khajanchi BK, Silver A, Graf J, Schein CH, Chopra AK, “Further characterization of a type III secretion system (T3SS) and of a new effector protein from a clinical isolate of Aeromonas hydrophila-Part I”, Microb Pathog. 2007 Oct; 43(4): 127-46, Epub 2007 May 18, PMID: 17644303 <http://www.ncbi.nlm.nih.gov/sites/entrez?Db=pubmed&Cmd=ShowDetailView&TermToSearch=17644303&ordinalpos=7&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum> | |||
< | 18. Johanna C Sierra , Giovanni Suarez , Jian Sha , Sheri M Foltz , Vsevolod L Popov , Cristi L Galindo , Harold R Garner , Ashok K Chopra “Biological characterization of a new type III secretion system effector from a clinical isolate of Aeromonas hydrophila-Part II”, Microb Pathog. 2007 Oct; 43(4): 147-60, Epub 2007 May 18, PMID: 17582731. <http://www.ncbi.nlm.nih.gov/sites/entrez?Db=pubmed&Cmd=ShowDetailView&TermToSearch=17582731&ordinalpos=12&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum> | ||
19. NCBI Taxonomy: Aeromonas hydrophila: | |||
<http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=644&lvl=3&lin=f&keep=1&srchmode=1&unlock> | |||
20. NCBI Genome Project: Aeromonas hydrophila subsp. hydrophila ATCC 7966: | |||
<http://www.ncbi.nlm.nih.gov/sites/entrez?Db=genomeprj&cmd=ShowDetailView&TermToSearch=16697? | |||
21. JGI IMG: <http://img.jgi.doe.gov/cgi-bin/pub/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=639633004> | |||
22. Northwest Fisheries Science Center <http://www.nwfsc.noaa.gov/> | |||
23. Wikipedia: Aeromonas hydrophila: <http://en.wikipedia.org/wiki/Aeromonas_hydrophila> | |||
24. Microbewiki: Aeromonas: <http://microbewiki.kenyon.edu/index.php/Aeromonas> | |||
25. Al-Fatlawy, H.N.K. and H.A. Al-Hadrawy. “Isolation and characterization of A. hydrophila from the Al-Jadryia river in Baghdad (Iraq)” American Journal of Educational Research, 2014, 2(8): 658-662. | |||
26. Harris, L.J., J.N. Farber, L.R. Beuchat, M.E. Parish, T.V. Suslow, E.H. Garrett, and F.F. Busta. | |||
“Outbreaks associated with fresh produce: incidence, growth, and survival of pathogens in fresh and fresh-cut produce” Comprehensive Reviews in Food Science and Food Safety, 2003, 2: 78-141. | |||
27. Janda, J.M. and S.L. Abbott. “The genus Aeromonas: taxonomy, pathogenicity, and infection” Clinical Microbiology Reviews, 2010, 23(1): 35-73. | |||
28. Radu, S., N. Ahmad, F.H. Ling, and A. Reezal. “Prevalence and resistance to antibiotics for Aeromonas species from retail fish in Malaysia” International Journal of Food Microbiology, 2003, 81:261-266. | |||
29. Abbott, S.L., W.K.W. Cheung, and J.M. Janda. "The genus Aeromonas: biochemical characteristics, atypical reactions, and phenotypic identification schemes" Journal of Clinical Microbiology, 2003, 41(6): 2348-2357. | |||
[ | Edited by student of [mailto:ralarsen@ucsd.edu Rachel Larsen] | ||
Latest revision as of 03:27, 9 May 2016
A Microbial Biorealm page on the genus Aeromonas Hydrophila
Classification
Higher order taxa
Cellular organism; Bacteria; Proteobacteria; Gammaproteobacteria; Aeromonadales; Aeromonadaceae; Aeromonas.
Species
|
NCBI: Taxonomy |
Aeromonas hydrophila
Aeromonas hydrophila is a complex of three species including A.hydrophila sensu stricto, A.bestiarum, and A. salmonicida. A.bestiarum, and A. salmonicida are rarely present in humans [27].
Subspecies: Aeromonas hydrophila subsp. anaerogenes, Aeromonas hydrophila subsp. decolorationis, Aeromonas hydrophila subsp. dhakensis, Aeromonas hydrophila subsp. hydrophila ATCC 7966, and Aeromonas hydrophila subsp. ranaei [19].
Description and significance
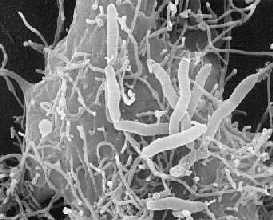
Aeromonas hydrophila is the most well known of the six species belonging to the genus Aeromonas. It is rod- shaped, non- spore forming, oxidase- positive, glucose- fermenting, facultative anaerobic, gram- negative bacterium that inhabits aquatic environments [1, 2]. Aeromonas hydrophila colonies on agar plates are smooth, convex, and rounded, and they are tan/buff-colored on trypticase soy agar [25, 29]. This bacterium can be found in fresh, brackish, estuarine, marine, chlorinated and unchlorinated water supplies worldwide, with highest numbers obtained in the warmer climates [1, 2, 3, 4]. Though most commonly found in aquatic environments, Aeromonas hydrophila can also inhabit soil [25, 28]. The bacterium has optimal growth at 28°C but can also grow at the limits from 4°C to 37°C [1]. Some strains of Aeromonas hydrophila are capable of causing disease in fish and amphibians as well as in humans who may acquire infections through open wounds or by ingestion of an adequate number of the organisms in water or food. Aeromonas hydrophila has been isolated from raw vegetables like carrots, lettuce, and parsley. No outbreaks of this A. hydrophila have been associated with raw vegetables yet, but it does have the ability to grow on them even at cold, refrigeration temperature [26]. This bacterium can digest materials such as gelatin, hemoglobin, and elastin. Aeromonas hydrophila was isolated from diseased cold- and warm- blooded animals for over 100 years and from humans since the early 1950s [1, 5]. It is also hard to kill because it is a resistant bacterium. Aeromonas hydrophila is resistant to chlorine and refrigeration or cold temperatures [24].
Genome structure
The complete genome of Aeromonas hydrophila strain ATCC 7966T was sequenced [14]. The genome is comprised of a single circular 4,744,448 bp chromosome with 61.5% GC content [20]. Its entire genome consists of 4,122 protein coding genes and 159 RNA genes: 128 tRNA genes amd 30 rRNA genes. It was possible to assign putative functions to 72.3% of the CDSs, while 21.5% possessed similarity to genes of unknown function, and no function could be proposed for 6.2% of the CDSs [21]. The complete genome sequence of Aeromonas hydrophila ATCC 7966T reveals mechanisms contributing to virulence and metabolic condition that allow the organism to grow in a variety of environments and explains how Aeromonas hydrophila is able to survive in polluted or oxygen- poor environments and to colonize and cause illness in humans and other hosts. However, two recognized virulence markers, a type III secretion system and a lateral flagellum, that are reported in other Aeromonas hydrophila strains are not identified in the sequenced isolate, ATCC 7966T. Given the ubiquity and free- living lifestyle of this organism, there is relatively little evidence of fluidity in terms of mobile elements in the genome of this particular strain. Thus, the Aeromonas hydrophila genome sequence provides valuable insights into its ability to flourish in both aquatic and host environments [14].
A number of virulence factors produced by Aeromonas species, however, their association with diarrhea have not been clearly linked [14]. Recently, a cytotoxic enterotoxin (Act), a heat- unstable cytotonic enterotoxin (Alt), and a heat- stable cytotonic enterotoxin (Ast) has been characterized from a diarrheal isolated of Aeromonas hydrophila. The Act is a single- chain polypeptide that is one of the primary genes to make this species pathogenic. Act is aerolysin related and has hemolytic, cytotoxic, and enterotoxic activities. Alt and Ast also have genes that contribute to its toxicity, but are not related to cholera toxin [6].
Cell structure and metabolism
Aeromonas hydrophila are gram- negative straight rods with rounded ends. They usually grow from 0.3 to 1.0 μm in diameter and 1.0 to 3.5 μm in length. Aeromonas hydrophila does not form endospores, and can grow in temperatures as low as 4°C. This bacterium is motile by polar flagella. Aeromonas hydrophila are heterotrophic organisms. They can exist in both aerobic and anaerobic environments, hence it is a facultative aerobe that ferments both glucose and sucrose( both A/-). It is oxidase positive. Aeromonas hydrophila can also digest gelatin, hemoglobin, and elastin [23, 24].
Aeromonas hydrophila has comprehensive biosynthetic abilities. The TCA cycle is complete and complemented by TCA cycle intermediates which can be replenished from acetate for use in various biosynthetic reactions. The Entner- Doudoroff and glycolytic pathways are intact, while the pentose phosphate pathway appears to be missing the oxidative branch. Complete multi- step pathways for synthesizing all amino acids are predicted, as are biosynthetic pathways for numerous cofactors such as biotin, glutathione, ubiquinone and menaquinone, pantothenate, thiamine, riboflavin, heme, molybdopterin, iron- sulfur clusters, coenzyme A, and tetrahydrofolate [14]. Aeromonas hydrophila has the oxygen- sensitive IscSUA- HscBA- Fdx system for the biosynthesis of iron- sulfur clusters rather than the oxygen- resistant SUF system, typically associated with aerobic and facultatively anaerobic organisms [14]. Under anaerobic growth conditions of Aeromonas hydrophila, a NiFe uptake hydrogenase may act as electron carrier to appropriate acceptors, and immediately downstream is a putative nickel transporter, followed by various hydrogenase maturations. Additionally, nitrate is converted to nitrite and then to ammonia in the general assimilatory pathway. Nitrate may also act as an electron acceptor for anaerobic respiration [14].
Sulfate assimilation is accomplished by reduction of sulfate to sulfide with the participation of a sulfate adenylyltransferase, adenylylsulfate kinase, and phosphoadenosine phosphosulfate reductase. The sulfite product from the assimilation pathway is reduced by sulfite reductase to sulfide for incorporation into an amino acid, peptide, protein, etc. Additionally, Aeromonas hydrophila has a dissimilatory anaerobic sulfite reductase involved in dissimilation of oxidized anions for energy transduction. This enzyme catalyzes hydrogen sulfide production from sulfite, which is regulated by electron acceptors from hydrogen or an organic substrate, and serves as an important energy- conserving step. It also confers the ability to synthesize cysteine anaerobically. Other electron acceptor sources utilized by Aeromonas hydrophila include tetrathionate, fumarate, and trimethyl- N- oxide [14].
Ecology
Aeromonas hydrophila are ubiquitous bacteria which are found in a variety of aquatic environments worldwide, including bottled water, chlorinated water, well water, and heavily polluted waters. The organism is posted in the Contaminant Candidate List by the Environmental Protection Agency, and U.S. water supplies are routinely examined for it. Even Aeromonas hydrophila confer the metabolic versatility to persist in its aquatic habitats or that facilitate ecological interactions with other prokaryotic and eukaryotic organisms [14]. Aeromonas hydrophila infections elevate occurrence with environmental changes, in contaminated environments, change in the temperature, and when an organism is already infected with a virus or another bacterium [23].
Pathology
Aeromonas hydrophila causes a variety of diseases in both fish and human populations. The ubiquitous nature of the bacterium in aquatic environments provides significant opportunity for animals, mainly fish and amphibians to contact and ingest organisms [14]. Aeromonas hydrophila is very toxic to many organisms because of its structure. When it enters the body of fish, amphibians, or humans, it travels via the bloodstream to the first available organ. It produces aerolysin cytotoxic enterotoxin (Act) which is one of the major virulence factors. Its toxin is produced and secreted by the cell from a type II secretion system. The toxin binds to high- affinity receptors and undergoes oligomerization to form a heptameric pore- forming complex which allows passage of small molecules in the plasma membrane, resulting in permeabilization of the cell, cell death, and eventually tissue destruction. Aeromonas hydrophila is also known as an opportunistic pathogenic bacterium, meaning they only infect hosts with weakened immune responses [6, 7]. Other virulence functions include a surface layer which inhibits complement- mediated killing, type IV pili for attachment, a set of extracellular proteases which can cause tissue damage, the ability to form capsules, and polar and lateral flagella [14, 25]. The polar flagella work to adhere this bacteria to surfaces, and the lateral flagella allow for colonization [25]. Aeromonas hydrophila also contains lipopolysaccharide on its outer membrane, which is a virulence factor in human infections [25]. Though Aeromonas hydrophila is considered a pathogenic bacterium, scientists have not been able to prove that it is the actual cause of some of the diseases it is associated with. It is believed that this bacterium aids in the infection of diseases, but do not cause the diseases themselves [14].
The disease in fish and amphibians
Aeromonas hydrophila cause illness mainly in fish and amphibians because this bacterium lives in aquatic environments. It is related to a disease found in frogs called red leg that causes internal or fatal hemorrhage. When infected with Aeromonas hydrophila, fish develop ulcers, fin rot, tail rot, and hemorrhagic septicaemia [12, 13]. Specially, hemorrhagic septicaemia causes lesions that lead to scale shedding, hemorrhages in the gills and anal area, ulcers, exophthalmia, and abdominal swelling [1, 23]. A study of retail fish in Malaysia conducted in 2003 had a total of 87 market fish samples. 11.5% of these samples contained Aeromonas hydrophila. They then looked at the antibiotic resistance patterns of the Aeromonas species in these fish and found that there was frequent resistance to ampicillin, carbenicillin, erythromycin, and streptomycin [28].
The disease in humans
Aeromonas hydrophila is also pathogenic to humans. It causes gastroenteritis which can affect anyone, but it most occurs in young children and people who have compromised immune systems or growth problems [12, 13]. This bacterium is linked to two types of gastroenteritis. The first type is a disease similar to cholera which causes rice- water diarrhea. Mild symptoms include fever and chills, but patients who become overwhelming bacterial infection with Aeromonas hydrophila often exhibit abdominal pain, nausea, vomiting, and diarrhea [1, 23]. The other type of disease is dysenteric gastroenteritis that causes loose stools filled with blood and mucus. Dysenteric gastroenteritis is the most severe out of the two types, and can last for multiple weeks. Aeromonas hydrophila is also associated with cellulitis, an infection that causes inflammation in the skin tissue. It also causes diseases such as myonecrosis and eczema in people with compromised immune systems [1, 14, 23].
Treatment
The most common treatment for Aeromonas hydrophila infection in humans are broad-spectrum antibiotics, like tetracycline [25]. It is particularly susceptible to fluoroquinolones (a family of antibiotics). The most effective were levofloxacin, gatifloxacin, ciprofloxacin, and moxifloxacin. Resistance to these antibiotics is rare [27].
Application to Biotechnology
Aeromonas hydrophila is a gram- negative opportunistic pathogen in fish, amphibians, and humans. Many bacterial pathogens of animals and plants have been shown to inject anti- host virulence determinants into the hosts by a type III secretion system (TTSS). Degenerate primers based on lcrD family genes are present in every known TTSS that are allowed us to locate the TTSS gene cluster in Aeromonas hydrophila AH- 1 [15]. A series of genome walking steps facilitate to identify 25 open reading frames that encode proteins homologous to TTSSs in other bacteria. With the PCR- based analysis, it shows that the presence of lcrD homologs in all of the 33 strains of Aeromonas hydrophila isolated from various sources. Insertional inactivation of two of the TTSS genes led to decreased cytotoxicity in carp epithelial cells, increased phagocytosis, and reduced virulence in blue gourami. These results indicate that a TTSS is required for Aeromonas hydrophila pathogenesis. The TTSS identified may facilitate to develop suitable vaccines as well as to understand more of the pathogenesis of Aeromonas hydrophila [15].
Current Research
Aeromonas hydrophila AH- 3 AexT is an ADP-ribosylating toxin secreted through the type III secretion system [16].
We cloned and sequenced an ADP- ribosylating toxin (AexT) from a mesophilic Aeromonas hydrophila strain AH-3 with a type III secretion system (T3SS). This toxin only showed homology, in genes and proteins, with the first half of Aeromonas salmonicida AexT. The Aeromonas hydrophila AexT showed ADP- ribosyltransferase activity, translocation through the T3SS system, and this Aeromonas hydrophila T3SS system is inducible under calcium- depleted conditions. The Aeromonas hydrophila aexT mutant showed a slight reduction in their virulence assayed by several methods when compared to the wild- type strain, while an Aeromonas hydrophila T3SS mutant is highly reduced in virulence on the same assays. The Aeromonas hydrophila AexT is the first described and the smallest T3SS effector toxin found in mesophilic Aeromonas with a functional T3SS [16].
Further characterization of a type III secretion system (T3SS) and of a new effector protein from a clinical isolate of Aeromonas hydrophila- Part I [17].
A type III secretion system (T3SS)- associated cytotoxin, AexT, with ADP-ribosyltransferase activity and homology to Pseudomonas aeruginosa bifuncational toxins ExoT/S, was recently identified from a fish pathogen Aeromonas salmonicida. In this study, we reported the molecular characterization of an aexT- like toxin gene (designated as aexU) from a diarrheal isolate SSU of Aeromonas hydrophila. The aexU gene was 1539 bp in length and encoded a protein of 512 amino acid residues. The NH(2)- terminus of AexU (aa residues 1-231) exhibited a 67% homology with the NH(2)-terminus of AexT from Aeromonas salmonicida. Importantly, its COOH- terminus (aa residues 232-512) had no homology with any known functional proteins in the database; however, the full-length AexU retained ADP- ribosyltransferase activity. The expression and subsequent secretion of AexU was T3SS dependent, as inactivation of the ascV gene that codes for an inner- membrane component of the T3SS channel from the wild- type bacterium, blocked translocation of AexU in HT-29 human colonic epithelial cells. We provided evidence that inactivation of acrV and axsE genes from Aeromonas hydrophila SSU, altered expression and/ or secretion of AexU. We deleted an aexU gene from the WT, as well as from the DeltaaopB mutant, of Aeromonas hydrophila, generating a single knockout (DeltaaexU) and a double knockout mutant, DeltaaopB/ DeltaaexU. Increased phagocytosis was observed in RAW264.7 murine macrophages infected with the DeltaaopB/ DeltaaexU mutant, as compared to macrophages when infected with the parental DeltaaopB strain. Further, mice infected with the DeltaaexU mutant had a 60% survival rate, compared to animals infected with the WT or the DeltaaexU- complemented strain that caused 90- 100% of the animals to die at a 2-3 LD(50s) dose. Immunization of mice with the recombinant AexU protected them from subsequent lethal challenge dose by the WT bacterium. Finally, we detected specific anti- AexU antibodies in the sera of mice that survived challenge by the WT bacterium, which may indicate that AexU plays an important role in the pathogenesis of Aeromonas infections [17].
Biological characterization of a new type III secretion system effector from a clinical isolate of Aeromonas hydrophila- Part II [18].
We recently identified a novel type III secretion system (T3SS) effector, AexU, from a diarrheal isolate SSU of Aeromonas hydrophila, and demonstrated that mice infected with the DeltaaexU mutant were significantly protected from mortality. Although the NH(2)- terminal domain of this toxin exhibits homology to AexT of Aeromonas salmonicida, a fish pathogen, and ExoT/ S of Pseudomonas aeruginosa, the COOH-terminal domain of AexU is unique, with no homology to any known proteins in the NCBI database. In this study, we purified the full- length AexU and its NH(2)- terminal (amino acid residues 1-231) and COOH-terminal (amino acid residues 232-512) domains after expression of their corresponding genes in E.coli as histidine- tag fusion proteins using the bacteriophage T7 RNA polymerase/ promoter-based pET-30a vector system. The full- length and NH(2)- and COOH-terminal domains of AexU exhibited ADP-ribosyltransferase activity, with the former two exhibiting much higher activity than the latter. These different forms of AexU were also successfully expressed and produced in the HeLa Tet- Off cell system using a pBI-EGFP vector, as demonstrated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, Western blot analysis, and intracellular staining of the toxin using flow cytometric analysis. Production of AexU in HeLa cells resulted in possible actin reorganization and cell rounding, as determined by phalloidin staining and confocal microscopy. Based on electron microscopy, the toxin also caused chromatin condensation, which is indicative of apoptosis. Apoptosis of HeLa cells expressing and producing AexU was confirmed by 7- amino actinomycin D (7-AAD) and MTT [3- (4,5- dimethylthiazol-2- yl)- 2,5- diphenyl tetrasodium bromide] assays, by detection of cytoplasmic histone-associated DNA fragments, and by activation of caspases 3 and 9. These effects were much more pronounced in host cells that expressed and produced the full-length or NH(2)- terminal domain of AexU, compared to those that expressed and produced the COOH- terminal domain or the vector alone. This study represents the first characterization of this novel T3SS effector [18].
References
1. Hayes, John. "Aeromonas hydrophila." Oregon State University. <http://hmsc.oregonstate.edu/classes/MB492/hydrophilahayes/>
2. Hazen, T.C., Flierman, C.B., Hirsch, R.P. and Esch, G.W, “Prevalence and distribution of Aeromonas hydrophila in the United States” Appl. Environ. Microbiol, 1978, 36:731–738.
3. Van der Kooj, D., “Properties of Aeromonads and their occurence and hygenic significance in drinking water”, Zentralb. Bakt. Hyg. B, 1988, 187:1–17.
4. Kaper, J.B., Lockman, H., Colwell, R.R. and Joseph, S.W., “Aeromonas hydrophila: ecology and toxigenicity of isolates form an estuary” J. Appl. Microbiol, 1980, 50:359–377.
5. Mathewson, J.J. and Dupont, H.L., “Aeromonas species: role as human pathogens”, In: Remington, J.S., Swartz, M.N. (eds.), “Current Clinical Topics in Infectious Diseases”, 1992, Vol12e. Cambridge: Blackwell Scientific. pp 26–36.
6. Albert, M. John et. al. "Prevalence of Enterotoxin Genes in Aeromonas spp. Isolated from Children with Diarrhea, Healthy Controls, and the Environment." Journal of Clinical Microbiology, October 2000, p. 3785-3790, Vol. 38, No. 10. < http://jcm.asm.org/cgi/content/full/38/10/3785>
7. Chopra, A.K., X.-J. Xu, D. Ribardo, M. Gonzalez, K. Kuhl, J. W. Peterson, and C. W. Houston. "The Cytotoxic Enterotoxin of Aeromonas hydrophila Induces Proinflammatory Cytokine Production and Activates Arachidonic Acid Metabolism in Macrophages." Infection and Immunity, May 2000, p. 2808-2818, Vol. 68, No.5. < http://iai.asm.org/cgi/content/full/68/5/2808>
8. Cipriano, Rocco L. "Aeromonas hydrophila and Motile Aereomonad Septicemias of Fish." Fish and Disease Leaflet 68. 2001. < http://www.lsc.usgs.gov/FHB/leaflets/FHB68.pdf >
9. Seetha KS, Jose BT, Jasthi A. "Meningitis due to aeromonas hydrophila." Indian J Med Microbiol 2004;22:191-192 < http://www.ijmm.org/article.asp?issn=0255-0857;year=2004;volume=22;issue=3;spage=191;epage=192;aulast=Seetha >
10. Xu, Xin-J., M. R. Ferguson, V. L. Popov, C. W. Houston, J. W. Peterson, and A. K. Chopra. "Role of a Cytotoxic Enterotoxin in Aeromonas-Mediated Infections: Development of Transposon and Isogenic Mutants." Infect Immun, August 1998, p. 3501-3509, Vol. 66, No. 8. < http://iai.asm.org/cgi/content/full/66/8/3501 >
11. Fulton, MacDonald. "THE BACTERIUM AEROMONAS HYDROPHILA FROM LIZARDS OF THE GENUS ANOLIS IN PUERTO RICO". Lousiana State University Medical Center, New Orleans, LA. <www.uprm.edu/publications/cjs/VOL05/P099-102.PDF>.
12. "Aeromonas hydrophila." Bad Bug Book Foodborne Pathogenic Microorganisms and Natural Toxins Handbook. <http://vm.cfsan.fda.gov/~mow/chap17.html>
13. "Aeromonas hydrophila and Related Bacteria." International Specialty Supply. <http://www.sproutnet.com/Reports/aeromonas_hydrophila.htm>
14. Rekha Seshadri, Sam W. Joseph, Ashok K. Chopra, Jian Sha, Jonathan Shaw, Joerg Graf, Daniel Haft, Martin Wu, Qinghu Ren, M. J. Rosovitz, Ramana Madupu, Luke Tallon, Mary Kim, Shaohua Jin, Hue Vuong, O. Colin Stine, Afsar Ali, Amy J. Horneman, and John F. Heidelberg “Genome Sequence of Aeromonas hydrophila ATCC 7966T: Jack of All Trades”, J Bacteriol. 2006 December. <http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1698176>
15. H. B. Yu, P. S. Srinivasa Rao, H. C. Lee, S. Vilches, S. Merino, J. M. Tomas, and K. Y. Leung, “A Type III Secretion System Is Required for Aeromonas hydrophila AH-1 Pathogenesis”, Infect Immun. 2004 March; 72(3): 1248–1256. <http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=356039>
16. Vilches S, Wilhelms M, Yu HB, Leung KY, Tomas JM, Merino S “Aeromonas hydrophila AH-3 AexT is an ADP-ribosylating toxin secreted through the type III secretion system”, Microb Pathog. 2007 Jul 1; PMID: 17689917 <http://www.ncbi.nlm.nih.gov/sites/entrez?Db=pubmed&Cmd=ShowDetailView&TermToSearch=17689917&ordinalpos=3&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum>
17. Sha J, Wang SF, Suarez G, Sierra JC, Fadl AA, Erova TE, Foltz SM, Khajanchi BK, Silver A, Graf J, Schein CH, Chopra AK, “Further characterization of a type III secretion system (T3SS) and of a new effector protein from a clinical isolate of Aeromonas hydrophila-Part I”, Microb Pathog. 2007 Oct; 43(4): 127-46, Epub 2007 May 18, PMID: 17644303 <http://www.ncbi.nlm.nih.gov/sites/entrez?Db=pubmed&Cmd=ShowDetailView&TermToSearch=17644303&ordinalpos=7&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum>
18. Johanna C Sierra , Giovanni Suarez , Jian Sha , Sheri M Foltz , Vsevolod L Popov , Cristi L Galindo , Harold R Garner , Ashok K Chopra “Biological characterization of a new type III secretion system effector from a clinical isolate of Aeromonas hydrophila-Part II”, Microb Pathog. 2007 Oct; 43(4): 147-60, Epub 2007 May 18, PMID: 17582731. <http://www.ncbi.nlm.nih.gov/sites/entrez?Db=pubmed&Cmd=ShowDetailView&TermToSearch=17582731&ordinalpos=12&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum>
19. NCBI Taxonomy: Aeromonas hydrophila: <http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=644&lvl=3&lin=f&keep=1&srchmode=1&unlock>
20. NCBI Genome Project: Aeromonas hydrophila subsp. hydrophila ATCC 7966: <http://www.ncbi.nlm.nih.gov/sites/entrez?Db=genomeprj&cmd=ShowDetailView&TermToSearch=16697?
21. JGI IMG: <http://img.jgi.doe.gov/cgi-bin/pub/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=639633004>
22. Northwest Fisheries Science Center <http://www.nwfsc.noaa.gov/>
23. Wikipedia: Aeromonas hydrophila: <http://en.wikipedia.org/wiki/Aeromonas_hydrophila>
24. Microbewiki: Aeromonas: <http://microbewiki.kenyon.edu/index.php/Aeromonas>
25. Al-Fatlawy, H.N.K. and H.A. Al-Hadrawy. “Isolation and characterization of A. hydrophila from the Al-Jadryia river in Baghdad (Iraq)” American Journal of Educational Research, 2014, 2(8): 658-662.
26. Harris, L.J., J.N. Farber, L.R. Beuchat, M.E. Parish, T.V. Suslow, E.H. Garrett, and F.F. Busta. “Outbreaks associated with fresh produce: incidence, growth, and survival of pathogens in fresh and fresh-cut produce” Comprehensive Reviews in Food Science and Food Safety, 2003, 2: 78-141.
27. Janda, J.M. and S.L. Abbott. “The genus Aeromonas: taxonomy, pathogenicity, and infection” Clinical Microbiology Reviews, 2010, 23(1): 35-73.
28. Radu, S., N. Ahmad, F.H. Ling, and A. Reezal. “Prevalence and resistance to antibiotics for Aeromonas species from retail fish in Malaysia” International Journal of Food Microbiology, 2003, 81:261-266.
29. Abbott, S.L., W.K.W. Cheung, and J.M. Janda. "The genus Aeromonas: biochemical characteristics, atypical reactions, and phenotypic identification schemes" Journal of Clinical Microbiology, 2003, 41(6): 2348-2357.
Edited by student of Rachel Larsen
