Marseilleviridae: Difference between revisions
No edit summary |
No edit summary |
||
| (9 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | {{Uncurated}} | ||
[[Image: | [[Image: Marseilleviridae_virion.jpg |thumb|300px|right|Legend. Image credit: Name or Publication.]] | ||
| Line 9: | Line 9: | ||
Viruses; Viridnavira; Bamfordviria; Nucleocytoviricota; megaviricetes [Others may be used. Use [http://www.ncbi.nlm.nih.gov/Taxonomy/ NCBI] link to find] | Viruses; Viridnavira; Bamfordviria; Nucleocytoviricota; megaviricetes; primascovirales [Others may be used. Use [http://www.ncbi.nlm.nih.gov/Taxonomy/ NCBI] link to find] | ||
| Line 25: | Line 25: | ||
'' | ''Marseillevirus marseillevirus'' | ||
| Line 31: | Line 31: | ||
==Description and Significance== | ==Description and Significance== | ||
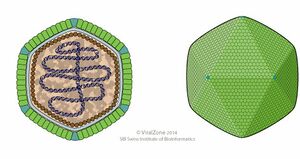
Marseilleviridae is the largest microbe discovered! It infects amoebae and the first one was isolated by culturing on amoebae from a water sample collected from a cooling tower in Paris, France. | |||
It has a capsid of around 250nm, with a 10nm thick capsid shell. It is roughly icosahedral in shape and surrounded by 12nm long fibers with globular ends. They are most common in freshwater, however two were found in humans (symptom free) and one was found in dipteran. | |||
marseilleviridae was detected in humans both with and without symptoms, and it was also found that there is a 30 day persistence of marseillevirus in rats and mice. these factors raise questions about the possible clinical significance of marseilleviruses. | |||
==Genome Structure== | ==Genome Structure== | ||
Like other viruses, the genome is a circular, double stranded DNA molecule. | |||
The genome is 368,454 base pairs in length with a CG content of 44.7% and encodes around 457 proteins. | |||
It is the 5th largest viral genome ever sequenced and is unusually rich in serine/threonine protein kinases. The prediction of 15 protein ligases suggests that versatile signaling is an extremely important aspect of the interaction between marseilleviridae and amoeba. | |||
Marseillevirus also encodes proteins which were not previously seen in nucleocytoplasmic large DNA viruses, specifically histone-like proteins. | |||
==Cell Structure, Metabolism and Life Cycle== | ==Cell Structure, Metabolism and Life Cycle== | ||
Phagocytosis by acanthamoeba is generally triggered by particles greater than 500nm. Marseilleviridae is 250nm in diameter, so therefore this is less likely to occur. | |||
In 2016, there was a study that found Marseilleviridae has different methods for penetrating amoeba. | |||
All Marseilleviruses have been reported to show traits for their replication and Verizon morphogenetic which is similar to Marseilleviridae Marseilleviridae. These steps occur in the cytoplasm and in viral factories, areas near the amoeba nucleus. The replication cycle takes 12 hours with the eclipse phase 2hours post infection. | |||
Incapacity of Virophage- lack of long fibrils and proteins R135 or homologs in Marseilleviridae might explain this incapacity, however further research is needed. | |||
==Ecology and Pathogenesis== | ==Ecology and Pathogenesis== | ||
Marseilleviridae is found in aquatic habitats. | |||
Marseilleviridae was detected in humans both with and without symptoms, and it was also found that there is a 30 day persistence of marseillevirus in rats and mice. these factors raise questions about the possible clinical significance of marseilleviruses. | |||
==References== | |||
[Sample reference] [http://ijs.sgmjournals.org/cgi/reprint/50/2/489 Takai, K., Sugai, A., Itoh, T., and Horikoshi, K. "''Palaeococcus ferrophilus'' gen. nov., sp. nov., a barophilic, hyperthermophilic archaeon from a deep-sea hydrothermal vent chimney". ''International Journal of Systematic and Evolutionary Microbiology''. 2000. Volume 50. p. 489-500.] | |||
Colson, P., Pagnier, I., Yoosuf, N., Fournous, G., La Scola, B., Raoult, D., 2012. “Marseilleviridae, a new family of giant viruses infecting amoebae”., springer link, https://link.springer.com/article/10.1007/s00705-012-1537-y [11.15.2022] | |||
Aherfi, s., La scola, B., Pagnier, I., Roult, D., Colson, P., 2014. “The expanding family Marseilleviridae”. Pubmed. https://pubmed.ncbi.nlm.nih.gov/25104553/ [11.15.2022] | |||
Boyer, M., Yutin, N., Pagnier, I., Barrassi., Fournous, G., Espinosa, L., Robert, C., Azza, S., Sun, S., Rossmann, M., Suzan-monti, M., La scola, B., Koonin, T., Raoult, D., 2009. “Giant Marseilleviridae highlights the role of amoebae as a melting pot in emergence of dimeric microorganisms”. Pubmed., https://pubmed.ncbi.nlm.nih.gov/20007369/ ., [11.15.2022] | |||
Salmi-bounsiar, D., Rolland, C., Aherfi S., Boudjemaa, H., leuasseur A., cascola, B., Colson, P., 2021., “Marseilleviruses, N update in 2021” ., https://www.frontiersin.org/articles/10.3389/fmicb.2021.648731/full#:~:text=.%2C%202020).-,Conclusion,of%20giant%20viruses%20of%20amoeba. ., [11.15.2022] | |||
==Author== | ==Author== | ||
Latest revision as of 20:18, 2 December 2022
Classification
Viruses; Viridnavira; Bamfordviria; Nucleocytoviricota; megaviricetes; primascovirales [Others may be used. Use NCBI link to find]
Species
|
NCBI: [1] |
Marseillevirus marseillevirus
Description and Significance
Marseilleviridae is the largest microbe discovered! It infects amoebae and the first one was isolated by culturing on amoebae from a water sample collected from a cooling tower in Paris, France. It has a capsid of around 250nm, with a 10nm thick capsid shell. It is roughly icosahedral in shape and surrounded by 12nm long fibers with globular ends. They are most common in freshwater, however two were found in humans (symptom free) and one was found in dipteran. marseilleviridae was detected in humans both with and without symptoms, and it was also found that there is a 30 day persistence of marseillevirus in rats and mice. these factors raise questions about the possible clinical significance of marseilleviruses.
Genome Structure
Like other viruses, the genome is a circular, double stranded DNA molecule. The genome is 368,454 base pairs in length with a CG content of 44.7% and encodes around 457 proteins. It is the 5th largest viral genome ever sequenced and is unusually rich in serine/threonine protein kinases. The prediction of 15 protein ligases suggests that versatile signaling is an extremely important aspect of the interaction between marseilleviridae and amoeba. Marseillevirus also encodes proteins which were not previously seen in nucleocytoplasmic large DNA viruses, specifically histone-like proteins.
Cell Structure, Metabolism and Life Cycle
Phagocytosis by acanthamoeba is generally triggered by particles greater than 500nm. Marseilleviridae is 250nm in diameter, so therefore this is less likely to occur. In 2016, there was a study that found Marseilleviridae has different methods for penetrating amoeba.
All Marseilleviruses have been reported to show traits for their replication and Verizon morphogenetic which is similar to Marseilleviridae Marseilleviridae. These steps occur in the cytoplasm and in viral factories, areas near the amoeba nucleus. The replication cycle takes 12 hours with the eclipse phase 2hours post infection.
Incapacity of Virophage- lack of long fibrils and proteins R135 or homologs in Marseilleviridae might explain this incapacity, however further research is needed.
Ecology and Pathogenesis
Marseilleviridae is found in aquatic habitats. Marseilleviridae was detected in humans both with and without symptoms, and it was also found that there is a 30 day persistence of marseillevirus in rats and mice. these factors raise questions about the possible clinical significance of marseilleviruses.
References
Colson, P., Pagnier, I., Yoosuf, N., Fournous, G., La Scola, B., Raoult, D., 2012. “Marseilleviridae, a new family of giant viruses infecting amoebae”., springer link, https://link.springer.com/article/10.1007/s00705-012-1537-y [11.15.2022]
Aherfi, s., La scola, B., Pagnier, I., Roult, D., Colson, P., 2014. “The expanding family Marseilleviridae”. Pubmed. https://pubmed.ncbi.nlm.nih.gov/25104553/ [11.15.2022]
Boyer, M., Yutin, N., Pagnier, I., Barrassi., Fournous, G., Espinosa, L., Robert, C., Azza, S., Sun, S., Rossmann, M., Suzan-monti, M., La scola, B., Koonin, T., Raoult, D., 2009. “Giant Marseilleviridae highlights the role of amoebae as a melting pot in emergence of dimeric microorganisms”. Pubmed., https://pubmed.ncbi.nlm.nih.gov/20007369/ ., [11.15.2022]
Salmi-bounsiar, D., Rolland, C., Aherfi S., Boudjemaa, H., leuasseur A., cascola, B., Colson, P., 2021., “Marseilleviruses, N update in 2021” ., https://www.frontiersin.org/articles/10.3389/fmicb.2021.648731/full#:~:text=.%2C%202020).-,Conclusion,of%20giant%20viruses%20of%20amoeba. ., [11.15.2022]
Author
Page authored by Abbigail McArthur, student of Prof. Bradley Tolar at UNC Wilmington.