Halorubrum sodomense: Difference between revisions
No edit summary |
|||
| (32 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | |||
==Classification== | ==Classification== | ||
| Line 10: | Line 10: | ||
"Halorubrum sodomense" | "Halorubrum sodomense" | ||
Discovered in 1980 | Discovered in 1980 (Oren, 1983) | ||
{| | {| | ||
| Line 19: | Line 19: | ||
|} | |} | ||
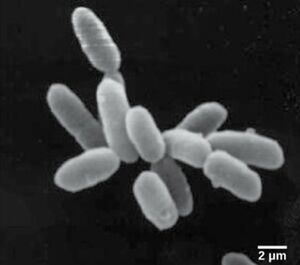
[[Image: Picture Halo.jpg|thumb|300px|right|"Haloruburm sodomense". | |||
Image credit: Bodaker, I, Itai, S, Suzuki, MT, Feingersch, R, Rosenberg, M, Maguire, ME, Shimshon, B, and others. Comparative community genomics in the Dead Sea: An increasingly extreme environment. The ISME Journal 4 (2010): 399–407, doi:10.1038/ismej.2009.141. published online 24 December 2009. ↵]] | |||
==Description and Significance== | ==Description and Significance== | ||
Grows in high salt concentrated water, Uses ATP synthesis from a sunlight driven photoreceptor protein, Important for assisting with development of optogenetics, Salt tolerant | Grows in high salt concentrated water, Uses ATP synthesis from a sunlight driven photoreceptor protein, Important for assisting with development of optogenetics, Salt tolerant (NCBI Taxonomy Browser) | ||
==Genome Structure== | ==Genome Structure== | ||
In the genome: Two chromosomes and one plasmid, Circular | In the genome: Two chromosomes and one plasmid, Circular shaped microbe (Oren, 1983), Proteins functioning in high saline/low temperature environments, Lives in hostile environments with immense solar exposure and highly concentrated seawater (Bodaker et al. 2010). | ||
==Cell Structure, Metabolism and Life Cycle== | ==Cell Structure, Metabolism and Life Cycle== | ||
On the surface: AR3 (Archaerhodopsin) retrieves energy from sun, Growth stems from high ion concentration of Mg2+, Create salt-tolerant bacteria | On the surface: AR3 (Archaerhodopsin) retrieves energy from sun (NCBI Taxonomy Browser), Growth of the microbe stems from high ion concentration of Mg2+, Create salt-tolerant bacteria, Lives in a 10x higher sodium concentrated environment, Presents an acidic level pH of 6.0 (Bodaker et al. 2010). | ||
==Ecology and Pathogenesis== | ==Ecology and Pathogenesis== | ||
Located on surface | Located on the surface of the Dead Sea and other high salt concentration bodies of water, Recombination has genetic information exchanged, mesophilic temperature ranges (BacDive), Ranks as a biosafety risk group level 1 (Admin, 2010) | ||
==References== | ==References== | ||
| Line 44: | Line 47: | ||
Bodaker, I, Itai, S, Suzuki, MT, Feingersch, R, Rosenberg, M, Maguire, ME, Shimshon, B, and others. Comparative community genomics in the Dead Sea: An increasingly extreme environment. The ISME Journal 4 (2010): 399–407, doi:10.1038/ismej.2009.141. published online 24 December 2009. ↵ | Bodaker, I, Itai, S, Suzuki, MT, Feingersch, R, Rosenberg, M, Maguire, ME, Shimshon, B, and others. Comparative community genomics in the Dead Sea: An increasingly extreme environment. The ISME Journal 4 (2010): 399–407, doi:10.1038/ismej.2009.141. published online 24 December 2009. ↵ | ||
Oren, Aharon. “Halobacterium Sodomense Sp. Nov., a Dead Sea Halobacterium with an Extremely High Magnesium Requirement.” International Journal of Systematic and Evolutionary Microbiology, Microbiology Society, 1 Apr. 1983, https://www.microbiologyresearch.org/content/journal/ijsem/10.1099/00207713-33-2-381. | |||
“Taxonomy Browser (Root).” National Center for Biotechnology Information, U.S. National Library of Medicine, https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi. | “Taxonomy Browser (Root).” National Center for Biotechnology Information, U.S. National Library of Medicine, https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi. | ||
Latest revision as of 19:39, 13 December 2022
Classification
Archaea; Euryarchaeota; Halobacteria; Haloferacales; Halorubraceae
Species
"Halorubrum sodomense"
Discovered in 1980 (Oren, 1983)
|
NCBI: [1] |
Description and Significance
Grows in high salt concentrated water, Uses ATP synthesis from a sunlight driven photoreceptor protein, Important for assisting with development of optogenetics, Salt tolerant (NCBI Taxonomy Browser)
Genome Structure
In the genome: Two chromosomes and one plasmid, Circular shaped microbe (Oren, 1983), Proteins functioning in high saline/low temperature environments, Lives in hostile environments with immense solar exposure and highly concentrated seawater (Bodaker et al. 2010).
Cell Structure, Metabolism and Life Cycle
On the surface: AR3 (Archaerhodopsin) retrieves energy from sun (NCBI Taxonomy Browser), Growth of the microbe stems from high ion concentration of Mg2+, Create salt-tolerant bacteria, Lives in a 10x higher sodium concentrated environment, Presents an acidic level pH of 6.0 (Bodaker et al. 2010).
Ecology and Pathogenesis
Located on the surface of the Dead Sea and other high salt concentration bodies of water, Recombination has genetic information exchanged, mesophilic temperature ranges (BacDive), Ranks as a biosafety risk group level 1 (Admin, 2010)
References
Admin. “Legislative Texts and Technical Rules - TRBA 466 Classification of Prokaryotes (Bacteria and Archaea) into Risk Groups.” BAuA, Dec. 2010, https://www.baua.de/EN/Service/Legislative-texts-and-technical-rules/Rules/TRBA/TRBA-466.html.
BacDive. “Halorubrum Sodomense RD 26 Is a Mesophilic Archaeon of the Family Halorubraceae.” BacDive, https://bacdive.dsmz.de/strain/5939.
Bodaker, I, Itai, S, Suzuki, MT, Feingersch, R, Rosenberg, M, Maguire, ME, Shimshon, B, and others. Comparative community genomics in the Dead Sea: An increasingly extreme environment. The ISME Journal 4 (2010): 399–407, doi:10.1038/ismej.2009.141. published online 24 December 2009. ↵
Oren, Aharon. “Halobacterium Sodomense Sp. Nov., a Dead Sea Halobacterium with an Extremely High Magnesium Requirement.” International Journal of Systematic and Evolutionary Microbiology, Microbiology Society, 1 Apr. 1983, https://www.microbiologyresearch.org/content/journal/ijsem/10.1099/00207713-33-2-381.
“Taxonomy Browser (Root).” National Center for Biotechnology Information, U.S. National Library of Medicine, https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi.
Author
Page authored by Hannah Arostegui, student of Prof. Bradley Tolar at UNC Wilmington.
