Nitrosospira lacus: Difference between revisions
No edit summary |
|||
| (26 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
==Higher order taxa== | ==Higher order taxa== | ||
Domain Bacteria | Domain Bacteria | ||
Phylum Proteobacteria | |||
Class Betaproteobacteria | |||
Order Nitrosomonadales | |||
Family nitrosomonadaceae | |||
Genus ''Nitrosospira'' | |||
==Species== | ==Species== | ||
Nitrosospira lacus | ''Nitrosospira lacus'' APG3<sup>T</sup> = NCIMB 14869<sup>T</sup> = LMG 27536<sup>T</sup> = ATCC BAA-2542<sup>T</sup> | ||
=Description and significance= | =Description and significance= | ||
Nitrosospira lacus is the 4th Nitrosospira species described (Urakawa et al. 2015). It is a sole representative of Cluster 0 Nitrosospira. | ''Nitrosospira lacus'' is the 4th ''Nitrosospira'' species described (Urakawa et al. 2015). It is a sole representative of Cluster 0 ''Nitrosospira''. APG3 was isolated from a shallow lake sediment in 2008 (Green Lake, WA, USA). Strain APG3 can grow at 4 °C but cannot grow at 35 °C, indicating that this bacterium is psychrotolerant. It should be noted that the strain is able to grow over a wide range of pH (pH 5–9), which is greater than the pH range of any previously reported ammonia-oxidizing bacteria in pure culture. However, the optimum pH is between 7.0 to 8.0, thus, APG3 is acid tolerant and not acidophilic. Some ammonia-oxidizing archaea show better adaptation to low pH soil environments (Lehtovirta-Morley et al. 2011). One another unique feature of this bacterium is that ''N. lacus'' is only ''Nitrosospira'' species that can grow at 4°C. Thus, this bacterium is psychrotolerant. | ||
=16S Ribosomal RNA Gene Information= | =16S Ribosomal RNA Gene Information= | ||
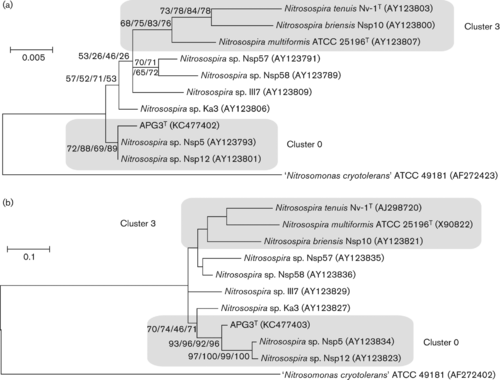
Phylogenetic analysis based on the 16S rRNA gene sequence showed that Nitrosospira multiformis (cluster 3) as the closest species with a validly published name (identity of 98.6 % to the type strain). | ''Nitrosospira lacus'' belongs to cluster 0 ''Nitrosospira''. It was documented as the first described species of cluster 0. Phylogenetic analysis based on the 16S rRNA gene sequence showed that ''Nitrosospira multiformis'' (cluster 3) as the closest species with a validly published name (identity of 98.6 % to the type strain). | ||
[[File:APG3 16S & amoA trees.gif|right|thumb|500px|Maximum-likelihood trees based on 16S rRNA (a) and ''amoA'' (b) gene sequences of members of the genus ''Nitrosospira''. Bars, 0.005 (a) and 0.1 (b) substitution units. Bootstrap values are listed as percentages at nodes (maximum-likelihood/neighbour-joining/maximum-parsimony/minimum-evolution) (Urakawa et al. 2015).]] | |||
=Genome Structure= | |||
The draft genome sequence comprises 3,107,181 bases, which is similar to the genome size of ''Nitrosospira multiformis'' ATCC 25196<sup>T</sup> (3.2 Mbp) but larger than the genomes of ''Nitrosomonas europaea'' ATCC 19718 (2.8 Mbp) and ''Nitrosomonas eutropha'' C71 (2.8 Mbp). The reported draft genome consists of 84 contigs with an average size of 41,181 bp. The DNA G+C content (53.6%) is similar to the G+C content of the genome of ''Nitrosospira multiformis'' ATCC 25196<sup>T</sup> (53.9%), but higher than that of ''Nitrosomonas europaea'' ATCC 19718 (50.7%) and ''Nitrosomonas eutropha'' C71 (48.5%). The draft genome contains 3147 protein-coding DNA sequences, 44 tRNA genes and a single 16S–23S–5S rRNA operon. | |||
The draft genome sequence comprises 3,107,181 bases, which is similar to the genome size of Nitrosospira multiformis ATCC | |||
=Cell structure and metabolism= | =Cell structure and metabolism= | ||
Strain APG3 grew optimally at 25 °C. It also grew at 4 and 10 °C, but no growth was observed at 35 °C. Growth of strain APG3 at pH 5 represents the most acidic conditions reported for AOB. However, this bacterium is not acidophilic because no growth was observed at pH 3 or 4 and the growth optimum was between pH 7 and 8. | Cells have a spiral shape, 0.8–1.2 µm wide and 1.3–1.7 µm long. APG3 is obligate chemolithotroph, and can oxidize ammonia to nitrite. It also uses urea. Cells have motility, and multiple genes involved in flagellum synthesis and function have been identified. Strain APG3 grew optimally at 25 °C. It also grew at 4 and 10 °C, but no growth was observed at 35 °C. Growth of strain APG3 at pH 5 represents the most acidic conditions reported for AOB. However, this bacterium is not acidophilic because no growth was observed at pH 3 or 4 and the growth optimum was between pH 7 and 8. | ||
=Ecology= | =Ecology= | ||
Strain APG3 was isolated from sandy sediment (30 cm depth) of Green Lake in North Central Seattle on 26 October 2008. Ammonia monooxygenase sequences (amoA) identified as N. lacus were found widely represented in samples from Asia (56 % of all sequences) and North America (43.1 %), but they were rather rare in samples from Europe (0.9 %). The distribution of APG3-like AOB is likely concentrated in the temperate climate zone, and limited in the tropical and subtropical zones. The psychrotolerant nature of APG3 reflects the global distribution of this novel species. No APG3-like AOB sequences have been reported from pelagic or coastal waters, including estuarine and tidal flats, indicating that this species originates on land. Moreover, there are no reports from wastewater treatment plants. Some sequences (6.1 % of the total) have been retrieved from soil environments (farm, forest, turf grass and acid sulfate soils), while the majority (93.9 % in total) were found in water-saturated or moisture-rich soils (e.g. freshwater sediment and paddy fields). | Strain APG3 was isolated from sandy sediment (30 cm depth) of Green Lake in North Central Seattle on 26 October 2008. Ammonia monooxygenase sequences (''amoA'') identified as ''N. lacus'' were found widely represented in samples from Asia (56 % of all sequences) and North America (43.1 %), but they were rather rare in samples from Europe (0.9 %). The distribution of APG3-like AOB is likely concentrated in the temperate climate zone, and limited in the tropical and subtropical zones. The psychrotolerant nature of APG3 reflects the global distribution of this novel species. No APG3-like AOB sequences have been reported from pelagic or coastal waters, including estuarine and tidal flats, indicating that this species originates on land. Moreover, there are no reports from wastewater treatment plants. Some sequences (6.1 % of the total) have been retrieved from soil environments (farm, forest, turf grass and acid sulfate soils), while the majority (93.9 % in total) were found in water-saturated or moisture-rich soils (e.g. freshwater sediment and paddy fields). | ||
=Current Research= | =Current Research= | ||
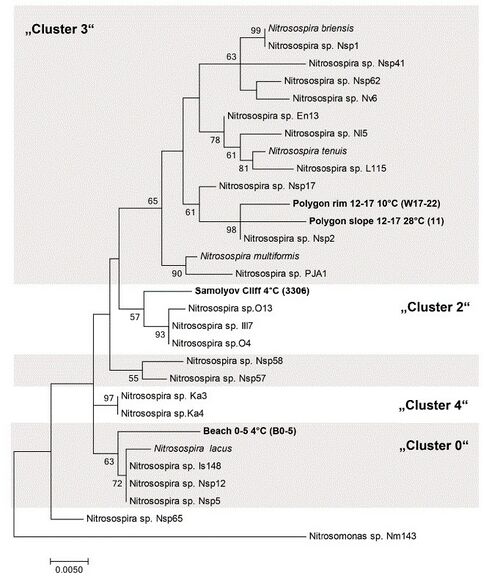
Cluster 0 ''Nitrosospira'' sequences were found in permafrost soil along with cluster 2 and 3 ''Nitrosospira'' (Sanders et al. 2019). In the tested environment, ammonia-oxidizing bacteria were more abundant than ammonia-oxidizing archaea. Not ''Nitrosomonas'' but ''Nitrosospira'' predominated. ''Nitrosospira'' may play a key role in ammonia-oxidation in permafrost soil environments. | |||
[[File:Sanders nitrosospira tree.jpg|right|thumb|500px|Maximum-likelihood 16S rRNA gene tree based on phylogenetic relationships between ''Nitrosospira'' enriched from arctic soil samples. Scale bar = 5% sequence divergence. Bootstrap values are shown.(Sanders et al. 2019).]] | |||
=References= | =References= | ||
Lehtovirta-Morley, L.E., Stoecker, K., Vilcinskas, A., Prosser, J.I. and Nicol, G.W., 2011. Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proceedings of the National Academy of Sciences, 108(38), pp.15892-15897. | |||
Sanders T, Fiencke C, Hüpeden J, Pfeiffer EM, Spieck E. Cold adapted ''Nitrosospira'' sp.: a potential crucial contributor of ammonia oxidation in cryosols of permafrost-affected landscapes in northeast Siberia. Microorganisms. 2019 Dec 14;7(12):699. | |||
Urakawa, H., Garcia, J.C., Nielsen, J.L., Le, V.Q., Kozlowski, J.A., Stein, L.Y., Lim, C.K., Pommerening-Roeser, A., Martens-Habbena, W., Stahl, D.A. and Klotz, M.G., 2015. ''Nitrosospira lacus'' sp. nov., a psychrotolerant, ammonia-oxidizing bacterium from sandy lake sediment. International journal of systematic and evolutionary microbiology, 65(Pt_1), pp.242-250. | |||
=Author= | =Author= | ||
Edited by Trevor Tubbs, student of Hidetoshi Urakawa in 2023 spring Microbial Ecology class (EVR4024C & EVR6907) at Florida Gulf Coast University | |||
Latest revision as of 17:04, 24 April 2023
Classification
Higher order taxa
Domain Bacteria Phylum Proteobacteria Class Betaproteobacteria Order Nitrosomonadales Family nitrosomonadaceae Genus Nitrosospira
Species
Nitrosospira lacus APG3T = NCIMB 14869T = LMG 27536T = ATCC BAA-2542T
Description and significance
Nitrosospira lacus is the 4th Nitrosospira species described (Urakawa et al. 2015). It is a sole representative of Cluster 0 Nitrosospira. APG3 was isolated from a shallow lake sediment in 2008 (Green Lake, WA, USA). Strain APG3 can grow at 4 °C but cannot grow at 35 °C, indicating that this bacterium is psychrotolerant. It should be noted that the strain is able to grow over a wide range of pH (pH 5–9), which is greater than the pH range of any previously reported ammonia-oxidizing bacteria in pure culture. However, the optimum pH is between 7.0 to 8.0, thus, APG3 is acid tolerant and not acidophilic. Some ammonia-oxidizing archaea show better adaptation to low pH soil environments (Lehtovirta-Morley et al. 2011). One another unique feature of this bacterium is that N. lacus is only Nitrosospira species that can grow at 4°C. Thus, this bacterium is psychrotolerant.
16S Ribosomal RNA Gene Information
Nitrosospira lacus belongs to cluster 0 Nitrosospira. It was documented as the first described species of cluster 0. Phylogenetic analysis based on the 16S rRNA gene sequence showed that Nitrosospira multiformis (cluster 3) as the closest species with a validly published name (identity of 98.6 % to the type strain).
Genome Structure
The draft genome sequence comprises 3,107,181 bases, which is similar to the genome size of Nitrosospira multiformis ATCC 25196T (3.2 Mbp) but larger than the genomes of Nitrosomonas europaea ATCC 19718 (2.8 Mbp) and Nitrosomonas eutropha C71 (2.8 Mbp). The reported draft genome consists of 84 contigs with an average size of 41,181 bp. The DNA G+C content (53.6%) is similar to the G+C content of the genome of Nitrosospira multiformis ATCC 25196T (53.9%), but higher than that of Nitrosomonas europaea ATCC 19718 (50.7%) and Nitrosomonas eutropha C71 (48.5%). The draft genome contains 3147 protein-coding DNA sequences, 44 tRNA genes and a single 16S–23S–5S rRNA operon.
Cell structure and metabolism
Cells have a spiral shape, 0.8–1.2 µm wide and 1.3–1.7 µm long. APG3 is obligate chemolithotroph, and can oxidize ammonia to nitrite. It also uses urea. Cells have motility, and multiple genes involved in flagellum synthesis and function have been identified. Strain APG3 grew optimally at 25 °C. It also grew at 4 and 10 °C, but no growth was observed at 35 °C. Growth of strain APG3 at pH 5 represents the most acidic conditions reported for AOB. However, this bacterium is not acidophilic because no growth was observed at pH 3 or 4 and the growth optimum was between pH 7 and 8.
Ecology
Strain APG3 was isolated from sandy sediment (30 cm depth) of Green Lake in North Central Seattle on 26 October 2008. Ammonia monooxygenase sequences (amoA) identified as N. lacus were found widely represented in samples from Asia (56 % of all sequences) and North America (43.1 %), but they were rather rare in samples from Europe (0.9 %). The distribution of APG3-like AOB is likely concentrated in the temperate climate zone, and limited in the tropical and subtropical zones. The psychrotolerant nature of APG3 reflects the global distribution of this novel species. No APG3-like AOB sequences have been reported from pelagic or coastal waters, including estuarine and tidal flats, indicating that this species originates on land. Moreover, there are no reports from wastewater treatment plants. Some sequences (6.1 % of the total) have been retrieved from soil environments (farm, forest, turf grass and acid sulfate soils), while the majority (93.9 % in total) were found in water-saturated or moisture-rich soils (e.g. freshwater sediment and paddy fields).
Current Research
Cluster 0 Nitrosospira sequences were found in permafrost soil along with cluster 2 and 3 Nitrosospira (Sanders et al. 2019). In the tested environment, ammonia-oxidizing bacteria were more abundant than ammonia-oxidizing archaea. Not Nitrosomonas but Nitrosospira predominated. Nitrosospira may play a key role in ammonia-oxidation in permafrost soil environments.
References
Lehtovirta-Morley, L.E., Stoecker, K., Vilcinskas, A., Prosser, J.I. and Nicol, G.W., 2011. Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proceedings of the National Academy of Sciences, 108(38), pp.15892-15897.
Sanders T, Fiencke C, Hüpeden J, Pfeiffer EM, Spieck E. Cold adapted Nitrosospira sp.: a potential crucial contributor of ammonia oxidation in cryosols of permafrost-affected landscapes in northeast Siberia. Microorganisms. 2019 Dec 14;7(12):699.
Urakawa, H., Garcia, J.C., Nielsen, J.L., Le, V.Q., Kozlowski, J.A., Stein, L.Y., Lim, C.K., Pommerening-Roeser, A., Martens-Habbena, W., Stahl, D.A. and Klotz, M.G., 2015. Nitrosospira lacus sp. nov., a psychrotolerant, ammonia-oxidizing bacterium from sandy lake sediment. International journal of systematic and evolutionary microbiology, 65(Pt_1), pp.242-250.
Author
Edited by Trevor Tubbs, student of Hidetoshi Urakawa in 2023 spring Microbial Ecology class (EVR4024C & EVR6907) at Florida Gulf Coast University