Methanosarcina barkeri: Difference between revisions
No edit summary |
|||
| (69 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
{{Curated}} | |||
{{Biorealm Genus}} | {{Biorealm Genus}} | ||
| Line 24: | Line 25: | ||
{| | {| | ||
| height="10" bgcolor="#FFDF95" | | | height="10" bgcolor="#FFDF95" | | ||
'''NCBI: [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=269797&lvl=3&p=mapview&p=has_linkout&p=blast_url&p=genome_blast&lin=f&keep=1&srchmode=1&unlock ''Methanosarcina barkeri str. | '''NCBI: [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=269797&lvl=3&p=mapview&p=has_linkout&p=blast_url&p=genome_blast&lin=f&keep=1&srchmode=1&unlock ''Methanosarcina barkeri str. fusaro'']''' | ||
|} | |} | ||
''Methanosarcina barkeri str. MS''<br /> | ''Methanosarcina barkeri str. MS''<br /> | ||
| Line 32: | Line 33: | ||
==Description and significance== | ==Description and significance== | ||
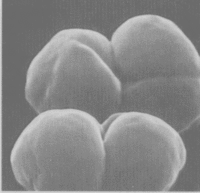
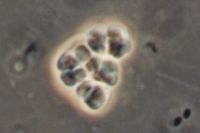
''Methansoarcina barkeri'' is | [[Image:Methanosarcina.jpg|thumb|Methanosarcina barkeri|200px|left|''Methanosarcina barkeri str. fusaro''. Photo: Dr. Steffen Battenberg. <br />Obtained from NCBI.]] | ||
<br /> | |||
''M. barkeri'' | The ''Methanosarcina'' are the only genus of the ''Methanosarcinaceae'' family, and of this, ''Methansoarcina barkeri'' is considered the representative species. Therefore, comments about ''M. barkeri'' can generally be applied to ''Methanosarcina''. (1) | ||
<br />''M. barkeri'' has been isolated from various places including sewage in Los Angeles and Urbana, Illinois. (1) The ''fusaro'' strain was isolated from mud samples taken from the freshwater lake Lago del Fusaro in Italy. It is also reported that ''M. barkeri'' lives in the rumen of cattle where it helps digest cellulose. (5,7) | |||
<br />''M. barkeri'' is the organism in which the 22nd amino acid, pyrrolysine, was discovered. Although this wasn't a totally revolutionary discovery because of the 21st amino acid's (selenocysteine) previous discovery, it is of particular interest because it is only found in archaea, particularly methanogens. Pyrrolysine plays a significant role in the process of methanogenesis. It should also be noted that pyrrolysine is the 21st amino acid that is directly charged to a tRNA. Selenocysteine is produced only after the tRNA has been charged with serine and that serine is modified into selenocysteine. (4) | |||
==Genome structure== | ==Genome structure== | ||
''Methanosarcina barkeri fusaro'' has one circular chromosome and one circular plasmid. | ''Methanosarcina barkeri str. fusaro'' was sequenced and annotated by the Joint Genome Institute at the University of Maryland and was funded by the Department of Energy. From that sequencing, it was determined that ''M. barkeri'' has one circular chromosome and one circular plasmid. (6) | ||
===Chromosome=== | ===Chromosome=== | ||
''M. barkeri's'' chromosome consists of 4,837,408 bp, fairly large for a methanogen. This is attributed to ''M. barkeri's'' ability to metabolize many different carbon sources. Of the 4.8Mbp, 69% of it codes for protein, yielding 3606 different proteins. There are also 73 structural RNAs, 132 pseudogenes, and various transposons. The genome is about 39% GC, relatively low. The chromosome sequence can be accessed with the GenBank ID CP000099 or with the Refseq ID NC_007355. (2,6) | |||
39% GC | |||
===Plasmid=== | |||
The plasmid consists of 36,358 bp, 58% of which are coding. The plasmid is only 33% GC. There were 26 genes identified on the plasmid with four genes thought to be associated with methanochondroitin synthesis, five putative transposases, and seven hypothetical genes whose function is unknown. The plasmid can be accessed with GenBank ID CP000098 or Refseq ID NC_007349. (2,6) | |||
GenBank | |||
Refseq | |||
<br />The plasmid is thought to replicated with the cell because it does not contain a site needed for replication via the rolling circle method, as does the plasmid of ''Methanosarcina acetivorans''. (2) | |||
<br />Through genome comparisons, it was found that none of the genes on the plasmid shared similarity with genes located on the plasmids of ''Methanosarcina acetivorans'' or ''Methanosarcina mazei''. (2) | |||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
'' | ''M. barkeri'' is non-motile, and contains no flagella, however, one species has been observed producing gas vesicles (see discussion below relating to gas vesicles). They have a tendency to form irregular clusters of cells, of various sizes, as the cells divide. The clusters can grow large enough to be seen by the naked eye. The cells are usually large and spherical and produce a positive Gram stain. (1) In liquid culture, the cells will form large zoogloeal masses. (3) The Gram-positive staining is likely a result of ''M. barkeri's'' thick cell wall (500nm), which is composed of an acid heteropolysaccharide. (1) The cell membrane is composed of relatively short lipids, consisting primarily of C25 hydrocarbons and C20 ethers. The majority of other methanogens consist of C30 hydrocarbons and a mixture of C20 and C40 ethers. (1) | ||
<br />There is some discussion amongst the community as to whether or not ''M. barkeri str. fusaro'' produces gas vesicles. Other species of ''Methanosarcina'' and one strain of ''M. barkeri'' have been shown to produce gas vesicles, possibly as a means of movement, but multiple studies have failed to produce gas vesicles in ''M. barkeri str. fusaro''. (8,2) This normally wouldn't lead to much discussion, however the genome of ''M. barkeri str. fusaro'' has the genes required for making gas vesicles, and therefore the strain should be able to make them. In the strain that did produce gas vesicles, it only did so in the presence of H2/CO2. It has been proposed that the gas vesicles are produced in response to hydrogen gradients. (2) | |||
<br />''M. barkeri'' is an extremely oxygen-sensitive anaerobic methanogen and unlike most methanogens which only use carbon dioxide, ''M. barkeri'' is able to ferment a variety of carbon sources including H2, methanol, methylamines, and acetate. (1,7) This offers a strong advantage to ''M. barkeri'' as it is immotile and therefore dependent on its environment for motility. Since it is able to utilize a variety of energy sources, it is relatively adaptable to its environment compared to its relatives. (2) | |||
==Ecology== | ==Ecology== | ||
''Methanosarcina barkeri'' is an anaerobe has been isolated from mud samples in lakes and bogs. ''M. barkeri'' also lives in the rumen of cows where it helps digest organic matter for the cow. A USA Today article | ''Methanosarcina barkeri'' is an anaerobe has been isolated from mud samples in lakes and bogs and sewage samples. ''M. barkeri'' also lives in the rumen of cows where it helps digest organic matter for the cow. A USA Today article (10) reported that up to 17% of the world's atmospheric methane comes from cows, a large majority of which would come from ''M. barkeri''. Because methane is a green house gas and can interfere with the ozone layer, this small organism may be partially responsible for two major environmental crises that we have face today: thinning of the ozone layer and global warming. (5,7,10) | ||
==Pathology== | ==Pathology== | ||
''Methanosarcina barkeri'' is an archaea and therefore | ''Methanosarcina barkeri'' is an archaea and therefore is not known to cause any diseases. | ||
==Application to Biotechnology== | ==Application to Biotechnology== | ||
As a | ''Methanosarcina barkeri'' is potentially useful to industry for two primary reasons. The first is related to its unique 22nd amino acid, pyrrolysine. Unlike the 21st amino acid, selenocysteine, pyrrolysine is only found and only needed in archaea. This opens up the possibility of using the tRNA of pyrrolysine to encode "unnatural" amino acids, allowing for advanced protein engineering. With the elucidation of the PYLIS (See [[#Current_Research | Current Research]]), scientists could insert these new amino acids into key locations in proteins to engineer novel functionality. (9) | ||
<br />The more obvious use of ''M. barkeri'' is to produce methane as an alternative fuel. As mentioned above, ''M. barkeri'' is able to use a variety of carbon sources for methanogenesis, so it is more flexible than other methanogens in terms of starting material. ''M. barkeri'' is also seemingly efficient. It has been said that a well-fed dairy cow can produce as much as 500L of intestinal gas in one day, 35% of which is methane. ''M. barkeri'' is thought to be primarily responsible for that 35%. (5) | |||
<br />Methane has an energy content of about 1000 BTU/cubic foot, which equals about 35 BTU/L. If ''M. barkeri'' produces only 20% of the methane passed by a dairy cow, which yields 100L of methane, enough to produce 3500BTU. This is enough energy to melt 24.5 pounds of ice or run a 1hp motor for 20 minutes. By this conservative estimate, ''M. barkeri'' holds enormous potential as an alternative energy supplier. (Wikipedia references: British Thermal Unit, Methane) | |||
==Current Research== | ==Current Research== | ||
[http://www.blackwell-synergy.com/doi/abs/10.1111/j.1365-2958.2006.05500.x Longstaff, DG and Blight, SK and Zhang, L and Green-Church, KB and Krzycki, JA. "In vivo contextual requirements for UAG translation as pyrrolysine". Molecular Microbiology. 2007. 63-1, p. 229-241.]<br /> | [http://www.blackwell-synergy.com/doi/abs/10.1111/j.1365-2958.2006.05500.x Longstaff, DG and Blight, SK and Zhang, L and Green-Church, KB and Krzycki, JA. "In vivo contextual requirements for UAG translation as pyrrolysine". Molecular Microbiology. 2007. 63-1, p. 229-241.]<br /> | ||
Pyrrolysine and selenocysteine are coded by a nonstandard translation of normal stop codons. For selenocysteine, it is known that | Pyrrolysine and selenocysteine are coded by a nonstandard translation of normal stop codons. For selenocysteine, it is known that the insertion of the amino acid, instead of message termination, is dependent on an insertion sequence that causes the formation of a secondary structure in the mRNA that favors insertion of selenocysteine. In this paper, the researchers demonstrate, experimentally, the requirement of a similar mechanism for pyrrolysine. This sequence, which they call the PYrroLysine Insertion Sequence (PYLIS), is located downstream and is not translated. The experiment consisted of the transfer of a pyrrolysine-containing gene from ''Methanosarcina barkeri'' to ''[[Methanosarcina acetivorans]]''. This is not a definitive study, as other mechanisms may contribute to the insertion of pyrrolysine or termination.<br /><br /> | ||
[http://www.sciencemag.org/cgi/content/abstract/315/5814/1003 Sato, T, Atomi, H, and Imanaka, T. "Archaeal Type III RuBisCOs Function in a Pathway for AMP Metabolism". Science. 2007. 315-5817, p. 1003.]<br /> | [http://www.sciencemag.org/cgi/content/abstract/315/5814/1003 Sato, T, Atomi, H, and Imanaka, T. "Archaeal Type III RuBisCOs Function in a Pathway for AMP Metabolism". Science. 2007. 315-5817, p. 1003.]<br /> | ||
Types I and II RuBisCO are found in all phototrophic organisms and are essential enzymes for the carbon fixation of the Calvin cycle. Type IV RuBisCO is found in ''Bacillus subtilis'' and is part of the Met salvage pathway. This paper describes the characterization of Type III RuBisCO, which is found only in select archaea, including ''Methanosarcina barkeri''. It was | Types I and II RuBisCO are found in all phototrophic organisms and are essential enzymes for the carbon fixation of the Calvin cycle. Type IV RuBisCO is found in ''[[Bacillus subtilis]]'' and is part of the Met salvage pathway. This paper describes the characterization of Type III RuBisCO, which is found only in select archaea, including ''Methanosarcina barkeri''. It was determined that Type III RuBisCO catalyzes the reaction of ribulose-1,5-bisphosphate to two molecules 3-phosphoglycerate, which can then enter either glycolysis or gluconeogenesis. This is mediated via two other enzymes (DeoA & E2b2) which break down AMP into the ribulose-1,5-bisphosphate.<br /><br /> | ||
[http://doi.wiley.com/10.1002/prot.21262 Chu, HM and Andrew, HJW. "Enzyme-Substrate Interactions Revealed by the Crystal Structures of the Archaeal Sulfolobus PTP-Fold Phosphatase and its Phosphopeptide Complexes". PROTEINS: Structure, Function, and Bioinformatics. 2007. 66, p. 996-1003.]<br /> | [http://doi.wiley.com/10.1002/prot.21262 Chu, HM and Andrew, HJW. "Enzyme-Substrate Interactions Revealed by the Crystal Structures of the Archaeal Sulfolobus PTP-Fold Phosphatase and its Phosphopeptide Complexes". PROTEINS: Structure, Function, and Bioinformatics. 2007. 66, p. 996-1003.]<br /> | ||
This paper describes the protein structure of a protein tyrosine phosphatase (PTP) that is important to signal transduction. This is of interest for those interested in ''Methanosarcina barkeri'' because protein phosphatases in archaea were first discovered in ''Methanosarcina barkeri''. This phosphatase was the methyltransferase activation protein, which is important for the activation of the methanol-using pathway of methanogenesis.<br /><br /> | |||
[http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=17360621&dopt=Citation Ambrogelly, A, Gundllapalli, S, Herring, S, Polycarpo, C, Frauer, C, and Soll, D. "Pyrrolysine is not hardwired for cotranslational insertion at UAG codons". Proceeds of the National Academy of Sciences. 2007. 104-9, p. 3141-3146.]<br /> | [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=17360621&dopt=Citation Ambrogelly, A, Gundllapalli, S, Herring, S, Polycarpo, C, Frauer, C, and Soll, D. "Pyrrolysine is not hardwired for cotranslational insertion at UAG codons". Proceeds of the National Academy of Sciences. 2007. 104-9, p. 3141-3146.]<br /> | ||
Pyrrolysine is encoded in ''Methanosarcinaceae'' via a naturally encoded tRNA that acts as a termination suppressor. In addition to the tRNA, a pyrrolysine-specific tRNA synthetase must also be present for pyrrolysine incorporation. The researchers in this paper transfered the Pyl tRNA synthetase to ''[[Escherichia coli]]'' and also transfered different mutants of the tRNA gene. From this, they were able to determine which positions of the tRNA are important for termination suppression based on assays performed. In addition to identifying positions of the tRNA that when mutated reduce suppression, they also found positions that increased suppression. <br /><br /> | |||
[http://www.nature.com/msb/journal/v2/n1/synopsis/msb4100046.html Feist, AM, Scholten, JCM, Palsson, BO, Brockman, FJ, and Ideker, T. "Modeling methanogenesis with a genome-scale metabolic reconstruction of Methanosarcina barkeri". Molecular Systems Biology. 2006. vol. 2-1.]<br /> | [http://www.nature.com/msb/journal/v2/n1/synopsis/msb4100046.html Feist, AM, Scholten, JCM, Palsson, BO, Brockman, FJ, and Ideker, T. "Modeling methanogenesis with a genome-scale metabolic reconstruction of Methanosarcina barkeri". Molecular Systems Biology. 2006. vol. 2-1.]<br /> | ||
The [http://systemsbiology.ucsd.edu systems biology] group at the University of California, San Diego reconstructed the metabolic network of ''Methanosarcina barkeri''. This involved determining reaction kinetics, stoichiometric coefficients, determining metabolic pathways, and determining protein interactions. The reconstruction for ''Methanosarcina barkeri'' is especially interesting because it will allow for the simulation of optimized methane-producing mutants as determined by metabolic engineering. | |||
==References== | ==References== | ||
[http://mmbr.asm.org/cgi/reprint/43/2/260.pdf Balch, WE, Fox, GE, Magrum, LJ, Woese, CR, and Wolfe, RS. "Methanogens: Reevaluation of a Unique Biological Group". ''Microbiological Reviews''. June 1979. p. 260-296. | [http://mmbr.asm.org/cgi/reprint/43/2/260.pdf (1)] Balch, WE, Fox, GE, Magrum, LJ, Woese, CR, and Wolfe, RS. "Methanogens: Reevaluation of a Unique Biological Group". ''Microbiological Reviews''. June 1979. p. 260-296. | ||
<br />[http://jb.asm.org/cgi/content/abstract/188/22/7922 Maeder DL, Anderson I, Brettin TS, Bruce DC, Gilna P, Han CS, Lapidus A, Metcalf WW, Saunders E, Tapia R, Sowers KR. "The Methanosarcina barkeri Genome: Comparative Analysis with | <br /><br />[http://jb.asm.org/cgi/content/abstract/188/22/7922 (2)] Maeder DL, Anderson I, Brettin TS, Bruce DC, Gilna P, Han CS, Lapidus A, Metcalf WW, Saunders E, Tapia R, Sowers KR. "The Methanosarcina barkeri Genome: Comparative Analysis with with Methanosarcina acetivorans and Methanosarcina mazei Reveals Extensive Rearrangement within Methanosarcinal Genomes". ''Journal of Bacteriology''. 2006. 188-22, p. 7922-7931. | ||
<br />[http://jb.asm.org/cgi/reprint/61/1/81.pdf Stadtman, TC and Barker, HA. "STUDIES ON THE METHANE FERMENTATION IX. The Origin of Methane in the Acetate and Methanol Fermentations by Methanosarcina". ''Journal of Bacteriology''. 1951. 61-1, p. 81-86.] | <br /><br />[http://jb.asm.org/cgi/reprint/61/1/81.pdf (3)] Stadtman, TC and Barker, HA. "STUDIES ON THE METHANE FERMENTATION IX. The Origin of Methane in the Acetate and Methanol Fermentations by Methanosarcina". ''Journal of Bacteriology''. 1951. 61-1, p. 81-86.] | ||
<br /> | <br /><br />[http://www.sciencemag.org/cgi/content/summary/sci;296/5572/1409 (4)] Atkins, JF and Gesteland, R. "The 22nd Amino Acid". Science. 2002. 296-5572, p. 1409-1410. | ||
<br />[http://www.sciencemag.org/cgi/content/summary/sci;296/5572/1409 Atkins, JF and Gesteland, R. "The 22nd Amino Acid". Science. 2002. 296-5572, p. 1409-1410. | <br /><br />[http://genome.jgi-psf.org/finished_microbes/metba/metba.home.html (5)] US Department of Energy Joint Genome Institute Organism Detail page for ''Methanosarcina barkeri'' | ||
<br /> | <br /><br />[http://cmr.tigr.org/tigr-scripts/CMR/GenomePage.cgi?org=ntmb02 (6)] TIGR Comprehensive Microbial Resource: ''Methanosarcina barkeri fusaro'' Genome Page | ||
<br />[http://genome.jgi-psf.org/finished_microbes/metba/metba.home.html US Department of Energy Joint Genome Institute Organism Detail page for ''Methanosarcina barkeri'' | <br /><br />[http://www.ebi.ac.uk/2can/genomes/archaea/Methanosarcina_barkeri.html (7)] EMBL-EBI: ''Methanosarcina barkeri'' | ||
<br />[http://cmr.tigr.org/tigr-scripts/CMR/GenomePage.cgi?org=ntmb02 TIGR Comprehensive Microbial Resource: ''Methanosarcina barkeri fusaro'' Genome Page] | <br /><br />[http://arjournals.annualreviews.org/doi/pdf/10.1146/annurev.mi.31.100177.001521?cookieSet=1 (8)] Mah, RA and Ward, DM and Baresi, L. and Glass, TL. "Biogenesis of Methane". "Annual Review of Microbiology". 1977. 31-1, p. 309-341. | ||
<br /><br />[http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=17360621&dopt=Citation (9)] Ambrogelly, A, Gundllapalli, S, Herring, S, Polycarpo, C, Frauer, C, and Soll, D. "Pyrrolysine is not hardwired for cotranslational insertion at UAG codons". Proceeds of the National Academy of Sciences. 2007. 104-9, p. 3141-3146. | |||
<br /><br />[http://www.accessmylibrary.com/coms2/summary_0286-25543388_ITM (10)] Cutting cattle's methane emissions. | |||
<hr> | |||
Edited by Ian Kerman, a student of [mailto:ralarsen@ucsd.edu Rachel Larsen] at [http://www.ucsd.edu UCSD]. | |||
Latest revision as of 15:10, 4 July 2011
A Microbial Biorealm page on the genus Methanosarcina barkeri
Classification
Higher order taxa
|
NCBI: Archaea : Euryarchaeota : Methanomicrobia : Methanosarcinales : Methanosarcinaceae : Methanosarcina |
Species
|
NCBI: Methanosarcina barkeri |
Strains
Methanosarcina barkeri str. MS
Methanosarcina barkeri str. W
Methanosarcina barkeri str. 227
Description and significance
The Methanosarcina are the only genus of the Methanosarcinaceae family, and of this, Methansoarcina barkeri is considered the representative species. Therefore, comments about M. barkeri can generally be applied to Methanosarcina. (1)
M. barkeri has been isolated from various places including sewage in Los Angeles and Urbana, Illinois. (1) The fusaro strain was isolated from mud samples taken from the freshwater lake Lago del Fusaro in Italy. It is also reported that M. barkeri lives in the rumen of cattle where it helps digest cellulose. (5,7)
M. barkeri is the organism in which the 22nd amino acid, pyrrolysine, was discovered. Although this wasn't a totally revolutionary discovery because of the 21st amino acid's (selenocysteine) previous discovery, it is of particular interest because it is only found in archaea, particularly methanogens. Pyrrolysine plays a significant role in the process of methanogenesis. It should also be noted that pyrrolysine is the 21st amino acid that is directly charged to a tRNA. Selenocysteine is produced only after the tRNA has been charged with serine and that serine is modified into selenocysteine. (4)
Genome structure
Methanosarcina barkeri str. fusaro was sequenced and annotated by the Joint Genome Institute at the University of Maryland and was funded by the Department of Energy. From that sequencing, it was determined that M. barkeri has one circular chromosome and one circular plasmid. (6)
Chromosome
M. barkeri's chromosome consists of 4,837,408 bp, fairly large for a methanogen. This is attributed to M. barkeri's ability to metabolize many different carbon sources. Of the 4.8Mbp, 69% of it codes for protein, yielding 3606 different proteins. There are also 73 structural RNAs, 132 pseudogenes, and various transposons. The genome is about 39% GC, relatively low. The chromosome sequence can be accessed with the GenBank ID CP000099 or with the Refseq ID NC_007355. (2,6)
Plasmid
The plasmid consists of 36,358 bp, 58% of which are coding. The plasmid is only 33% GC. There were 26 genes identified on the plasmid with four genes thought to be associated with methanochondroitin synthesis, five putative transposases, and seven hypothetical genes whose function is unknown. The plasmid can be accessed with GenBank ID CP000098 or Refseq ID NC_007349. (2,6)
The plasmid is thought to replicated with the cell because it does not contain a site needed for replication via the rolling circle method, as does the plasmid of Methanosarcina acetivorans. (2)
Through genome comparisons, it was found that none of the genes on the plasmid shared similarity with genes located on the plasmids of Methanosarcina acetivorans or Methanosarcina mazei. (2)
Cell structure and metabolism
M. barkeri is non-motile, and contains no flagella, however, one species has been observed producing gas vesicles (see discussion below relating to gas vesicles). They have a tendency to form irregular clusters of cells, of various sizes, as the cells divide. The clusters can grow large enough to be seen by the naked eye. The cells are usually large and spherical and produce a positive Gram stain. (1) In liquid culture, the cells will form large zoogloeal masses. (3) The Gram-positive staining is likely a result of M. barkeri's thick cell wall (500nm), which is composed of an acid heteropolysaccharide. (1) The cell membrane is composed of relatively short lipids, consisting primarily of C25 hydrocarbons and C20 ethers. The majority of other methanogens consist of C30 hydrocarbons and a mixture of C20 and C40 ethers. (1)
There is some discussion amongst the community as to whether or not M. barkeri str. fusaro produces gas vesicles. Other species of Methanosarcina and one strain of M. barkeri have been shown to produce gas vesicles, possibly as a means of movement, but multiple studies have failed to produce gas vesicles in M. barkeri str. fusaro. (8,2) This normally wouldn't lead to much discussion, however the genome of M. barkeri str. fusaro has the genes required for making gas vesicles, and therefore the strain should be able to make them. In the strain that did produce gas vesicles, it only did so in the presence of H2/CO2. It has been proposed that the gas vesicles are produced in response to hydrogen gradients. (2)
M. barkeri is an extremely oxygen-sensitive anaerobic methanogen and unlike most methanogens which only use carbon dioxide, M. barkeri is able to ferment a variety of carbon sources including H2, methanol, methylamines, and acetate. (1,7) This offers a strong advantage to M. barkeri as it is immotile and therefore dependent on its environment for motility. Since it is able to utilize a variety of energy sources, it is relatively adaptable to its environment compared to its relatives. (2)
Ecology
Methanosarcina barkeri is an anaerobe has been isolated from mud samples in lakes and bogs and sewage samples. M. barkeri also lives in the rumen of cows where it helps digest organic matter for the cow. A USA Today article (10) reported that up to 17% of the world's atmospheric methane comes from cows, a large majority of which would come from M. barkeri. Because methane is a green house gas and can interfere with the ozone layer, this small organism may be partially responsible for two major environmental crises that we have face today: thinning of the ozone layer and global warming. (5,7,10)
Pathology
Methanosarcina barkeri is an archaea and therefore is not known to cause any diseases.
Application to Biotechnology
Methanosarcina barkeri is potentially useful to industry for two primary reasons. The first is related to its unique 22nd amino acid, pyrrolysine. Unlike the 21st amino acid, selenocysteine, pyrrolysine is only found and only needed in archaea. This opens up the possibility of using the tRNA of pyrrolysine to encode "unnatural" amino acids, allowing for advanced protein engineering. With the elucidation of the PYLIS (See Current Research), scientists could insert these new amino acids into key locations in proteins to engineer novel functionality. (9)
The more obvious use of M. barkeri is to produce methane as an alternative fuel. As mentioned above, M. barkeri is able to use a variety of carbon sources for methanogenesis, so it is more flexible than other methanogens in terms of starting material. M. barkeri is also seemingly efficient. It has been said that a well-fed dairy cow can produce as much as 500L of intestinal gas in one day, 35% of which is methane. M. barkeri is thought to be primarily responsible for that 35%. (5)
Methane has an energy content of about 1000 BTU/cubic foot, which equals about 35 BTU/L. If M. barkeri produces only 20% of the methane passed by a dairy cow, which yields 100L of methane, enough to produce 3500BTU. This is enough energy to melt 24.5 pounds of ice or run a 1hp motor for 20 minutes. By this conservative estimate, M. barkeri holds enormous potential as an alternative energy supplier. (Wikipedia references: British Thermal Unit, Methane)
Current Research
Longstaff, DG and Blight, SK and Zhang, L and Green-Church, KB and Krzycki, JA. "In vivo contextual requirements for UAG translation as pyrrolysine". Molecular Microbiology. 2007. 63-1, p. 229-241.
Pyrrolysine and selenocysteine are coded by a nonstandard translation of normal stop codons. For selenocysteine, it is known that the insertion of the amino acid, instead of message termination, is dependent on an insertion sequence that causes the formation of a secondary structure in the mRNA that favors insertion of selenocysteine. In this paper, the researchers demonstrate, experimentally, the requirement of a similar mechanism for pyrrolysine. This sequence, which they call the PYrroLysine Insertion Sequence (PYLIS), is located downstream and is not translated. The experiment consisted of the transfer of a pyrrolysine-containing gene from Methanosarcina barkeri to Methanosarcina acetivorans. This is not a definitive study, as other mechanisms may contribute to the insertion of pyrrolysine or termination.
Sato, T, Atomi, H, and Imanaka, T. "Archaeal Type III RuBisCOs Function in a Pathway for AMP Metabolism". Science. 2007. 315-5817, p. 1003.
Types I and II RuBisCO are found in all phototrophic organisms and are essential enzymes for the carbon fixation of the Calvin cycle. Type IV RuBisCO is found in Bacillus subtilis and is part of the Met salvage pathway. This paper describes the characterization of Type III RuBisCO, which is found only in select archaea, including Methanosarcina barkeri. It was determined that Type III RuBisCO catalyzes the reaction of ribulose-1,5-bisphosphate to two molecules 3-phosphoglycerate, which can then enter either glycolysis or gluconeogenesis. This is mediated via two other enzymes (DeoA & E2b2) which break down AMP into the ribulose-1,5-bisphosphate.
Chu, HM and Andrew, HJW. "Enzyme-Substrate Interactions Revealed by the Crystal Structures of the Archaeal Sulfolobus PTP-Fold Phosphatase and its Phosphopeptide Complexes". PROTEINS: Structure, Function, and Bioinformatics. 2007. 66, p. 996-1003.
This paper describes the protein structure of a protein tyrosine phosphatase (PTP) that is important to signal transduction. This is of interest for those interested in Methanosarcina barkeri because protein phosphatases in archaea were first discovered in Methanosarcina barkeri. This phosphatase was the methyltransferase activation protein, which is important for the activation of the methanol-using pathway of methanogenesis.
Ambrogelly, A, Gundllapalli, S, Herring, S, Polycarpo, C, Frauer, C, and Soll, D. "Pyrrolysine is not hardwired for cotranslational insertion at UAG codons". Proceeds of the National Academy of Sciences. 2007. 104-9, p. 3141-3146.
Pyrrolysine is encoded in Methanosarcinaceae via a naturally encoded tRNA that acts as a termination suppressor. In addition to the tRNA, a pyrrolysine-specific tRNA synthetase must also be present for pyrrolysine incorporation. The researchers in this paper transfered the Pyl tRNA synthetase to Escherichia coli and also transfered different mutants of the tRNA gene. From this, they were able to determine which positions of the tRNA are important for termination suppression based on assays performed. In addition to identifying positions of the tRNA that when mutated reduce suppression, they also found positions that increased suppression.
Feist, AM, Scholten, JCM, Palsson, BO, Brockman, FJ, and Ideker, T. "Modeling methanogenesis with a genome-scale metabolic reconstruction of Methanosarcina barkeri". Molecular Systems Biology. 2006. vol. 2-1.
The systems biology group at the University of California, San Diego reconstructed the metabolic network of Methanosarcina barkeri. This involved determining reaction kinetics, stoichiometric coefficients, determining metabolic pathways, and determining protein interactions. The reconstruction for Methanosarcina barkeri is especially interesting because it will allow for the simulation of optimized methane-producing mutants as determined by metabolic engineering.
References
(1) Balch, WE, Fox, GE, Magrum, LJ, Woese, CR, and Wolfe, RS. "Methanogens: Reevaluation of a Unique Biological Group". Microbiological Reviews. June 1979. p. 260-296.
(2) Maeder DL, Anderson I, Brettin TS, Bruce DC, Gilna P, Han CS, Lapidus A, Metcalf WW, Saunders E, Tapia R, Sowers KR. "The Methanosarcina barkeri Genome: Comparative Analysis with with Methanosarcina acetivorans and Methanosarcina mazei Reveals Extensive Rearrangement within Methanosarcinal Genomes". Journal of Bacteriology. 2006. 188-22, p. 7922-7931.
(3) Stadtman, TC and Barker, HA. "STUDIES ON THE METHANE FERMENTATION IX. The Origin of Methane in the Acetate and Methanol Fermentations by Methanosarcina". Journal of Bacteriology. 1951. 61-1, p. 81-86.]
(4) Atkins, JF and Gesteland, R. "The 22nd Amino Acid". Science. 2002. 296-5572, p. 1409-1410.
(5) US Department of Energy Joint Genome Institute Organism Detail page for Methanosarcina barkeri
(6) TIGR Comprehensive Microbial Resource: Methanosarcina barkeri fusaro Genome Page
(7) EMBL-EBI: Methanosarcina barkeri
(8) Mah, RA and Ward, DM and Baresi, L. and Glass, TL. "Biogenesis of Methane". "Annual Review of Microbiology". 1977. 31-1, p. 309-341.
(9) Ambrogelly, A, Gundllapalli, S, Herring, S, Polycarpo, C, Frauer, C, and Soll, D. "Pyrrolysine is not hardwired for cotranslational insertion at UAG codons". Proceeds of the National Academy of Sciences. 2007. 104-9, p. 3141-3146.
(10) Cutting cattle's methane emissions.
Edited by Ian Kerman, a student of Rachel Larsen at UCSD.