Geosiphon-Nostoc Endosymbiosis: Difference between revisions
mNo edit summary |
|||
| (11 intermediate revisions by the same user not shown) | |||
| Line 27: | Line 27: | ||
''Geosiphon pyriformis'' is a fungus in the class Glomeromycetes, these fungi usually form arbuscular mycorrhizal symbiotic relationships with plants and cannot live without a symbiotic partner. ''G. pyriformis'' is the only fungus known form an endosymbiotic relationship with nitrogen-fixing cyanobacteria (''Nostoc punctiforme''). Microbiologists studying this relationship say that ''G. pyriformis'' represents an ideal candidate to investigate the origin of AMS (arbuscular mycorrhizal symbiosis) and the emergence of a unique endosymbiosis (Malar et al., 2021). | ''Geosiphon pyriformis'' is a fungus in the class Glomeromycetes, these fungi usually form arbuscular mycorrhizal symbiotic relationships with plants and cannot live without a symbiotic partner. ''G. pyriformis'' is the only fungus known form an endosymbiotic relationship with nitrogen-fixing cyanobacteria (''Nostoc punctiforme''). Microbiologists studying this relationship say that ''G. pyriformis'' represents an ideal candidate to investigate the origin of AMS (arbuscular mycorrhizal symbiosis) and the emergence of a unique endosymbiosis (Malar et al., 2021). | ||
= | =Genome Information= | ||
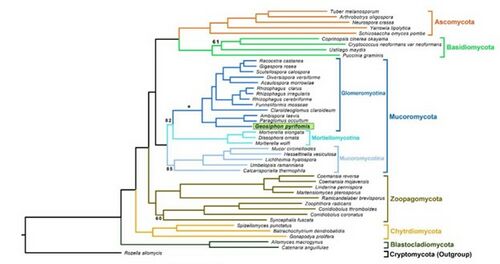
[[File:Geosiphon tree.jpg|thumbnail|500px|''Geosiphon pyriformis'' phylogenetic tree (Malar et al., 2021)]] | [[File:Geosiphon tree.jpg|thumbnail|500px|''Geosiphon pyriformis'' phylogenetic tree (Malar et al., 2021)]] | ||
The genome of ''N. punctiforme'' is 9.5 Mb, 41.5% GC, 7432 open reading frames, determined by shotgun sequencing | The genome of ''N. punctiforme'' is 9.5 Mb, 41.5% GC, with 7432 open reading frames, determined by shotgun sequencing. 45% of the genes found encode proteins with known or likely functions and 29% of which are unique to the species. More than 400 genes encoding sensor protein kinases were found which explains the species' ability to respond to environmental signals. (Meeks et al., 2001). | ||
The '' | The ''G. pyriformis'' genome sequence and modeling was performed by Nicolas Corradi and Mathu Malar at the University of Ottawa. It was determined that the genome was 128.32 Mbp with 24,195 genes, and had a GC% of 29.25. These scientists found that, in the genome, 365 genes represented putative secreted proteins, and 27% of those were candidate effectors (chemicals that aid in the initiation and maintenance of symbiosis). The researchers also looked for evidence of horizontal gene transfer between the two species and found 18 genes with bacterial potential and two protein encoding genes with significant sequence conservation with ''Nostoc'' or Gammaproteobacteria homologs (Malar et al., 2021). | ||
=Ecology and Symbiosis= | =Ecology and Symbiosis= | ||
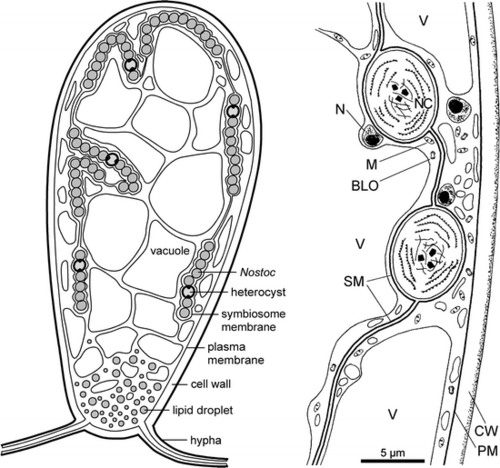
[[File:geopyr symbiosis.png|thumbnail|500px|Geosiphon-Nostoc symbiosis (Schubler et al., 2012)]] | [[File:geopyr symbiosis.png|thumbnail|500px|Geosiphon-Nostoc symbiosis (Schubler et al., 2012)]] | ||
As opposed to other AMF, which penetrate the plant cells to form arbuscules, ''G. pyriformis'' forms long fungal cells (‘bladders’) that enclose cyanobacteria, these bladder membranes are an extension of the fungal plasma membrane. Once the fungi have trapped the cyanobacteria in their bladders, the fungus supplies the cyanobacteria with water and inorganic nutrients while the cyanobacterium remains photosynthetically active and retains its ability to fix nitrogen using heterocysts. Findings also suggested that “during the switch from regular AMS to a fungal-cyanobacteria symbiosis, ''G. pyriformis'' evolved novel strategies to utilize lipids from its new host through the expansion of specific gene motifs. Specifically, the ''G. pyriformis'' genome carries a striking over-representation of lipase 3 protein domains that hydrolyze ester linkages of fatty acids. As ''Nostoc spp | As opposed to other AMF, which penetrate the plant cells to form arbuscules, ''G. pyriformis'' forms long fungal cells (‘bladders’) that enclose cyanobacteria, these bladder membranes are an extension of the fungal plasma membrane. Once the fungi have trapped the cyanobacteria in their bladders, the fungus supplies the cyanobacteria with water and inorganic nutrients while the cyanobacterium remains photosynthetically active and retains its ability to fix nitrogen using heterocysts. Findings also suggested that “during the switch from regular AMS to a fungal-cyanobacteria symbiosis, ''G. pyriformis'' evolved novel strategies to utilize lipids from its new host through the expansion of specific gene motifs. Specifically, the ''G. pyriformis'' genome carries a striking over-representation of lipase 3 protein domains that hydrolyze ester linkages of fatty acids. As ''Nostoc spp''. are known to produce a wide variety of extracellular lipids in high amounts, it is possible that these abundant lipids are released in the environment (like many other cellular compounds released by cyanobacteria) and are then broken down by lipases to be used as an energy resource by ''G. pyriformis''.”- (Malar et al. 2021). | ||
=References= | =References= | ||
Malar C, M., Krüger, M., Krüger, C., Wang, Y., Stajich, J. E., Keller, J., Chen, E. C. H., Yildirir, G., Villeneuve-Laroche, M., Roux, C., Delaux, P.-M., & Corradi, N. (2021). The genome of Geosiphon pyriformis reveals ancestral traits linked to the emergence of the arbuscular mycorrhizal symbiosis. Current Biology, 31(7). https://doi.org/10.1016/j.cub.2021.01.058 | Malar C, M., Krüger, M., Krüger, C., Wang, Y., Stajich, J. E., Keller, J., Chen, E. C. H., Yildirir, G., Villeneuve-Laroche, M., Roux, C., Delaux, P.-M., & Corradi, N. (2021). The genome of ''Geosiphon pyriformis'' reveals ancestral traits linked to the emergence of the arbuscular mycorrhizal symbiosis. Current Biology, 31(7). https://doi.org/10.1016/j.cub.2021.01.058 | ||
Schüßler, A. (2012). | Meeks, J. C. (2001). An overview of the genome of ''Nostoc punctiforme'', a multicellular, symbiotic cyanobacterium. Photosynthesis Research. https://doi.org/10.2172/841015 | ||
Schüßler, A. (2012). The Geosiphon–nostoc endosymbiosis and its role as a model for arbuscular mycorrhiza research. Fungal Associations, 77–91. https://doi.org/10.1007/978-3-642-30826-0_5 | |||
=Author= | =Author= | ||
Logan Meadors | Logan Meadors, a student of Dr. Hidetoshi Urakawa of Florida Gulf Coast University for Microbial Ecology EVR 4024C spring 2023. | ||
Latest revision as of 19:19, 23 March 2024
Classification
Higher order taxa
Domain: Bacteria, Phylum: Cyanobacteria, Class: Cyanophyceae, Order: Nostocales, Family: Nostocaceae, Genus: Nostoc
Domain: Fungi, Phylum: Mucoromycota, Class: Glomeromycetes, Order: Archaeosporales, Family: Geosiphonaceae, Genus: Geosiphon
Species
Nostoc punctiforme (ATTC 29133) Geosiphon pyriformis (NCBI 50956)
Description and significance
Nostoc punctiforme is a freshwater filamentous cyanobacterium that can be photoautotrophic, diazotrophic, heterotrophic, and/or symbiotic. It is broadly symbiotic, forming relationships with fungi, bryophytes, gymnosperms, and angiosperms. Its genome contains a large number of unique genes that likely play essential roles in cell differentiation and symbiosis (Meeks et al., 2001).
Geosiphon pyriformis is a fungus in the class Glomeromycetes, these fungi usually form arbuscular mycorrhizal symbiotic relationships with plants and cannot live without a symbiotic partner. G. pyriformis is the only fungus known form an endosymbiotic relationship with nitrogen-fixing cyanobacteria (Nostoc punctiforme). Microbiologists studying this relationship say that G. pyriformis represents an ideal candidate to investigate the origin of AMS (arbuscular mycorrhizal symbiosis) and the emergence of a unique endosymbiosis (Malar et al., 2021).
Genome Information
The genome of N. punctiforme is 9.5 Mb, 41.5% GC, with 7432 open reading frames, determined by shotgun sequencing. 45% of the genes found encode proteins with known or likely functions and 29% of which are unique to the species. More than 400 genes encoding sensor protein kinases were found which explains the species' ability to respond to environmental signals. (Meeks et al., 2001).
The G. pyriformis genome sequence and modeling was performed by Nicolas Corradi and Mathu Malar at the University of Ottawa. It was determined that the genome was 128.32 Mbp with 24,195 genes, and had a GC% of 29.25. These scientists found that, in the genome, 365 genes represented putative secreted proteins, and 27% of those were candidate effectors (chemicals that aid in the initiation and maintenance of symbiosis). The researchers also looked for evidence of horizontal gene transfer between the two species and found 18 genes with bacterial potential and two protein encoding genes with significant sequence conservation with Nostoc or Gammaproteobacteria homologs (Malar et al., 2021).
Ecology and Symbiosis
As opposed to other AMF, which penetrate the plant cells to form arbuscules, G. pyriformis forms long fungal cells (‘bladders’) that enclose cyanobacteria, these bladder membranes are an extension of the fungal plasma membrane. Once the fungi have trapped the cyanobacteria in their bladders, the fungus supplies the cyanobacteria with water and inorganic nutrients while the cyanobacterium remains photosynthetically active and retains its ability to fix nitrogen using heterocysts. Findings also suggested that “during the switch from regular AMS to a fungal-cyanobacteria symbiosis, G. pyriformis evolved novel strategies to utilize lipids from its new host through the expansion of specific gene motifs. Specifically, the G. pyriformis genome carries a striking over-representation of lipase 3 protein domains that hydrolyze ester linkages of fatty acids. As Nostoc spp. are known to produce a wide variety of extracellular lipids in high amounts, it is possible that these abundant lipids are released in the environment (like many other cellular compounds released by cyanobacteria) and are then broken down by lipases to be used as an energy resource by G. pyriformis.”- (Malar et al. 2021).
References
Malar C, M., Krüger, M., Krüger, C., Wang, Y., Stajich, J. E., Keller, J., Chen, E. C. H., Yildirir, G., Villeneuve-Laroche, M., Roux, C., Delaux, P.-M., & Corradi, N. (2021). The genome of Geosiphon pyriformis reveals ancestral traits linked to the emergence of the arbuscular mycorrhizal symbiosis. Current Biology, 31(7). https://doi.org/10.1016/j.cub.2021.01.058
Meeks, J. C. (2001). An overview of the genome of Nostoc punctiforme, a multicellular, symbiotic cyanobacterium. Photosynthesis Research. https://doi.org/10.2172/841015
Schüßler, A. (2012). The Geosiphon–nostoc endosymbiosis and its role as a model for arbuscular mycorrhiza research. Fungal Associations, 77–91. https://doi.org/10.1007/978-3-642-30826-0_5
Author
Logan Meadors, a student of Dr. Hidetoshi Urakawa of Florida Gulf Coast University for Microbial Ecology EVR 4024C spring 2023.