User:Pruner1: Difference between revisions
No edit summary |
No edit summary Tag: Manual revert |
||
| (25 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
==Introduction== | ==Introduction== | ||
Escherichia coli (<i>E. coli</i>) is a type of gram-negative, rod-shaped bacteria that has been used as a model organism in biology since it was discovered in 1885. The bacteria is named after German pediatrician Theodor Escherich, who discovered it in the stool of infants while looking for the cause of neonatal dysentery. | Escherichia coli (<i>E. coli</i>) is a type of gram-negative, rod-shaped bacteria that has been used as a model organism in biology since it was discovered in 1885. The bacteria is named after German pediatrician Theodor Escherich, who discovered it in the stool of infants while looking for the cause of neonatal dysentery. <ref name = "Lederberg Paper" > Lederberg, Joshua. 2004. E. Coli K-12. Microbiology Today, Volume 31, pg. 116. https://microbiologysociety.org/static/uploaded/f7395484-cf6e-40f2-9583e8d4e80ab18f.pdf</ref> <i>E. coli</i> has been put to good use since its discovery in 1885; discoveries involving <i>E. coli</i> have received eleven Nobel prizes and the bacteria has been used in countless experiments, making it one of the most important organisms in science. <i>E. coli</i> became one of the foremost model organisms owing to the fact that it is small, reproduces quickly, and can be grown and cultured easily. As a model organism, it has shaped knowledge in the fields of genetics and biology. Five groundbreaking experiments involving <i>E. coli</i> have been summarized below. | ||
==Experiment 1== | ==Experiment 1== | ||
Discovery of Genetic Code: | Discovery of Genetic Code: [[Image: Geneticcode.jpg|thumb|200px|right|Figure 1. The finished table of the genetic code. "The genetic code," by OpenStax College, Biology. | ||
In 1961, Marshall Nirenberg and Heinrich Matthaei made the groundbreaking discovery of the genetic code. While scientists understood that DNA served as a template for RNA and proteins, they didn’t fully understand how. Nirenberg and Matthaei mixed ribosomes, tRNA, and aminoacyl-tRNA synthetases from the <i>E. coli</i> with a synthetic RNA chain of uracil bases. <ref name= | . [https: http://cnx.org/contents/GFy_h8cu@9.87:QEibhJMi@8/The-Genetic-Code].]] | ||
In 1961, Marshall Nirenberg and Heinrich Matthaei made the groundbreaking discovery of the genetic code. While scientists understood that DNA served as a template for RNA and proteins, they didn’t fully understand how. Nirenberg and Matthaei mixed ribosomes, tRNA, and aminoacyl-tRNA synthetases from the <i>E. coli</i> with a synthetic RNA chain of uracil bases. <ref name = "Genetic Code" >Marshall Nirenberg. The genetic code. Nobel Lecture, December 12, 1968. (Accessed December 8, 2024). https://www.nobelprize.org/uploads/2018/06/nirenberg-lecture.pdf</ref> Essentially, they put all the tools of protein synthesis in a test tube and created their own RNA to see what proteins might be created from it. When mixed, the <i>E. coli</i> components read the chain of uracil bases and created a protein chain of only phenylalanine. The conclusion they drew was that uracil coded for the protein phenylalanine. Having broken into one example of the genetic code, the duo then fed different sequences of RNA through the elements to see what proteins were created. This laid the foundation of discovering how DNA translated RNA into proteins. It allowed scientists to understand how the 64 combinations of RNA codons, based on three nucleotides each, can encode the twenty standard amino acids. These combinations make up the translation we now call ‘genetic code’. <ref name = "ACS" > American Chemical Society National Historic Chemical Landmarks. Deciphering the Genetic Code. http://www.acs.org/content/acs/en/education/whatischemistry/landmarks/geneticcode.html (accessed December 8, 2024).</ref> Because the proteins, such as ribosomes, could be easily harvested from the <i>E. coli</i> and used <i>in vitro</i> in the cell, <i>E. coli</i> once again proved itself as an effective model organism. | |||
==Experiment 2== | ==Experiment 2== | ||
Discovery of semi-conservative replication: | |||
After Watson and Crick proved that DNA is shaped as a double helix, the next question many researchers had was how DNA replicated. There were three possible explanations for DNA replication: dispersive, semi-conservative, and conservative replication. Each of these theories are illustrated in the figure below. The dispersive theory hypothesized that the original parent strand and new daughter strand would each be broken up into several chunks and re-synthesized in a DNA strand patchwork of parent-daughter DNA. The semi-conservative theory hypothesized that one strand of parent DNA and one strand of daughter DNA would combine with each other to form the DNA double helix. <ref name = "Meselson-Stahl" >Geraldine Abir Am. The Meselson-Stahl Experiment. Published 15 May 2024. https://doi.org/10.1002/9780470015902.a0025093</ref> The conservative replication theory hypothesized that as DNA replicated, the parent strand would remain completely intact but serve as a template for the new daughter DNA strands. Thus, there would be one completely new daughter double-helix and the original parent double-helix. Meselson and Stahl used density-gradient centrifugation, a technique invented by Meselson, to settle this debate. The pair originally centrifuged DNA from a bacteriophage to try to figure out which replication theory was correct. However, the bacteriophage DNA broke apart during centrifugation and also replicated too quickly for the researchers to be able to tell which theory was correct. As a remedy, they turned to using trusty (<i>E. coli</i>) DNA. | |||
< | [[Image: Microbe wiki dna.jpg|thumb|500px|right|Figure 1. Illustration of the three leading theories on DNA replication. Howell-Moroney, Madeleine, "The Meselson-Stahl Experiment". Embryo Project Encyclopedia ( 2022-01-25 ). https://hdl.handle.net/10776/13325.]] | ||
< | researchers marked the parent strand of (<i>E. coli</i>) DNA with the isotope <sup>15</sup>N and put <sup>14</sup>N isotopes in the mixture surrounding the DNA. <ref name = "Meselson and Stahl" >Hernandez, Victoria. The Meselson-Stahl Experiment (1957–1958), by Matthew Meselson and Franklin Stahl. Published 04/08/2017</ref> By weighing the density of the DNA strands, which now varied because of the isotopes, they could see which isotopes the new daughter double-helix contained and use that knowledge to elucidate what method of DNA replication was occurring. If the DNA contained both <sup>15</sup>N and <sup>14</sup>N isotopes, the parent and daughter strands did combine, disproving the conservative hypothesis. Whirling the (<i>E. coli</i>) DNA strands around on the order of 14,000 times the force of gravity, the denser DNA was pushed towards the bottom while the lighter strands floated towards the top. The result showed a band of ½ <sup>14</sup>N strand DNA and ½ <sup>15</sup>N isotope-labeled DNA. These bands with a mix of the two isotopes disproved conservative DNA replication theory. Researching further, Meselson and Stalk measured the density of the new DNA strands, The density of these strands was the exact density in between <sup>15</sup>N and <sup>14</sup>N strands of DNA. If DNA followed dispersive replication theory, the weight of the strands would have varied slightly. Each DNA strand being the same weight proved the semiconservative model of DNA replication. Thus, (<i>E. coli</i>) contributed to the knowledge of DNA replication because of its tightly bonded DNA and well-timed cycles of replication. | ||
< | |||
< | |||
< | |||
< | |||
< | |||
< | |||
< | |||
< | |||
< | |||
==Experiment 3== | |||
Over 75,000 generations of <i>E. coli</i> have been used in Richard Lenski’s experiment on long-term evolution. <ref name = "Lenski Experiment" > Lenski, Richard. Convergence and Divergence in a Long-Term Experiment with Bacteria. The American Naturalist, Volume 190. August 2017. https://doi.org/10.1086/691209</ref> The Long Term Evolution Experiment (LTEE) is designed to explore questions about phenotypic and genotyping evolution, via the medium of <i>E. coli</i>. Beginning in 1988, the experiment has continued for over twenty years, and is one of the most convincing pieces of evidence towards the theory of evolution. Richard Lenski began this experiment by isolating twelve identical populations of <i>E. coli</i> on a medium of citrate, all of which came from the same ancestral source of <i>E. coli</i>. He fed each microbial community with a limited amount of glucose. From this set-up, Lenski took samples of the <i>E. coli</i> every day, and with the <i>E. coli</i> reproducing up to 7 times every day, he very quickly watched thousands of generations of <i>E. coli</i> evolve. One of the novel traits that <i>E. coli</i> evolved was the ability to use citrate as a source of energy. While at the beginning the <i>E. coli</i> could not use citrate, with the evolution of the Cit<sup>+</sup> trait in one of the populations. The Cit<sup>+</sup> mutation, as well as several others in the populations, serve as evidence that evolution occurs. This finding gave proof to what Darwin and the entire field of biology had long suspected: that populations can evolve. The Lenski experiment is considered a keystone in the field of modern day biology. It is revolutionary in the fields of genetics and biology. | |||
<ref name=" | |||
==References== | ==References== | ||
<references /> | <references /> | ||
<br>Edited by [ | <br>Edited by [Claire Pruner], student of Joan Slonczewski for BIOL 116, 2024, [http://www.kenyon.edu/index.xml Kenyon College]. | ||
<!--Do not edit or remove this line-->[[Category:Pages edited by students of Joan Slonczewski at Kenyon College]] | <!--Do not edit or remove this line-->[[Category:Pages edited by students of Joan Slonczewski at Kenyon College]] | ||
Latest revision as of 16:10, 13 December 2024
Introduction
Escherichia coli (E. coli) is a type of gram-negative, rod-shaped bacteria that has been used as a model organism in biology since it was discovered in 1885. The bacteria is named after German pediatrician Theodor Escherich, who discovered it in the stool of infants while looking for the cause of neonatal dysentery. [1] E. coli has been put to good use since its discovery in 1885; discoveries involving E. coli have received eleven Nobel prizes and the bacteria has been used in countless experiments, making it one of the most important organisms in science. E. coli became one of the foremost model organisms owing to the fact that it is small, reproduces quickly, and can be grown and cultured easily. As a model organism, it has shaped knowledge in the fields of genetics and biology. Five groundbreaking experiments involving E. coli have been summarized below.
Experiment 1
Discovery of Genetic Code:
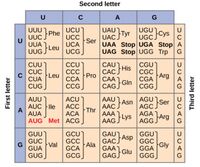
In 1961, Marshall Nirenberg and Heinrich Matthaei made the groundbreaking discovery of the genetic code. While scientists understood that DNA served as a template for RNA and proteins, they didn’t fully understand how. Nirenberg and Matthaei mixed ribosomes, tRNA, and aminoacyl-tRNA synthetases from the E. coli with a synthetic RNA chain of uracil bases. [2] Essentially, they put all the tools of protein synthesis in a test tube and created their own RNA to see what proteins might be created from it. When mixed, the E. coli components read the chain of uracil bases and created a protein chain of only phenylalanine. The conclusion they drew was that uracil coded for the protein phenylalanine. Having broken into one example of the genetic code, the duo then fed different sequences of RNA through the elements to see what proteins were created. This laid the foundation of discovering how DNA translated RNA into proteins. It allowed scientists to understand how the 64 combinations of RNA codons, based on three nucleotides each, can encode the twenty standard amino acids. These combinations make up the translation we now call ‘genetic code’. [3] Because the proteins, such as ribosomes, could be easily harvested from the E. coli and used in vitro in the cell, E. coli once again proved itself as an effective model organism.
Experiment 2
Discovery of semi-conservative replication:
After Watson and Crick proved that DNA is shaped as a double helix, the next question many researchers had was how DNA replicated. There were three possible explanations for DNA replication: dispersive, semi-conservative, and conservative replication. Each of these theories are illustrated in the figure below. The dispersive theory hypothesized that the original parent strand and new daughter strand would each be broken up into several chunks and re-synthesized in a DNA strand patchwork of parent-daughter DNA. The semi-conservative theory hypothesized that one strand of parent DNA and one strand of daughter DNA would combine with each other to form the DNA double helix. [4] The conservative replication theory hypothesized that as DNA replicated, the parent strand would remain completely intact but serve as a template for the new daughter DNA strands. Thus, there would be one completely new daughter double-helix and the original parent double-helix. Meselson and Stahl used density-gradient centrifugation, a technique invented by Meselson, to settle this debate. The pair originally centrifuged DNA from a bacteriophage to try to figure out which replication theory was correct. However, the bacteriophage DNA broke apart during centrifugation and also replicated too quickly for the researchers to be able to tell which theory was correct. As a remedy, they turned to using trusty (E. coli) DNA.
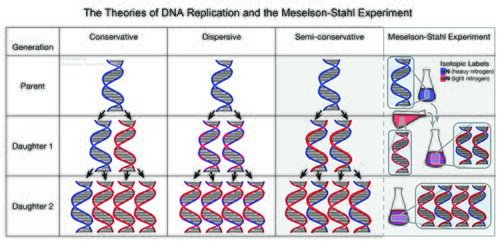
researchers marked the parent strand of (E. coli) DNA with the isotope 15N and put 14N isotopes in the mixture surrounding the DNA. [5] By weighing the density of the DNA strands, which now varied because of the isotopes, they could see which isotopes the new daughter double-helix contained and use that knowledge to elucidate what method of DNA replication was occurring. If the DNA contained both 15N and 14N isotopes, the parent and daughter strands did combine, disproving the conservative hypothesis. Whirling the (E. coli) DNA strands around on the order of 14,000 times the force of gravity, the denser DNA was pushed towards the bottom while the lighter strands floated towards the top. The result showed a band of ½ 14N strand DNA and ½ 15N isotope-labeled DNA. These bands with a mix of the two isotopes disproved conservative DNA replication theory. Researching further, Meselson and Stalk measured the density of the new DNA strands, The density of these strands was the exact density in between 15N and 14N strands of DNA. If DNA followed dispersive replication theory, the weight of the strands would have varied slightly. Each DNA strand being the same weight proved the semiconservative model of DNA replication. Thus, (E. coli) contributed to the knowledge of DNA replication because of its tightly bonded DNA and well-timed cycles of replication.
Experiment 3
Over 75,000 generations of E. coli have been used in Richard Lenski’s experiment on long-term evolution. [6] The Long Term Evolution Experiment (LTEE) is designed to explore questions about phenotypic and genotyping evolution, via the medium of E. coli. Beginning in 1988, the experiment has continued for over twenty years, and is one of the most convincing pieces of evidence towards the theory of evolution. Richard Lenski began this experiment by isolating twelve identical populations of E. coli on a medium of citrate, all of which came from the same ancestral source of E. coli. He fed each microbial community with a limited amount of glucose. From this set-up, Lenski took samples of the E. coli every day, and with the E. coli reproducing up to 7 times every day, he very quickly watched thousands of generations of E. coli evolve. One of the novel traits that E. coli evolved was the ability to use citrate as a source of energy. While at the beginning the E. coli could not use citrate, with the evolution of the Cit+ trait in one of the populations. The Cit+ mutation, as well as several others in the populations, serve as evidence that evolution occurs. This finding gave proof to what Darwin and the entire field of biology had long suspected: that populations can evolve. The Lenski experiment is considered a keystone in the field of modern day biology. It is revolutionary in the fields of genetics and biology.
References
- ↑ Lederberg, Joshua. 2004. E. Coli K-12. Microbiology Today, Volume 31, pg. 116. https://microbiologysociety.org/static/uploaded/f7395484-cf6e-40f2-9583e8d4e80ab18f.pdf
- ↑ Marshall Nirenberg. The genetic code. Nobel Lecture, December 12, 1968. (Accessed December 8, 2024). https://www.nobelprize.org/uploads/2018/06/nirenberg-lecture.pdf
- ↑ American Chemical Society National Historic Chemical Landmarks. Deciphering the Genetic Code. http://www.acs.org/content/acs/en/education/whatischemistry/landmarks/geneticcode.html (accessed December 8, 2024).
- ↑ Geraldine Abir Am. The Meselson-Stahl Experiment. Published 15 May 2024. https://doi.org/10.1002/9780470015902.a0025093
- ↑ Hernandez, Victoria. The Meselson-Stahl Experiment (1957–1958), by Matthew Meselson and Franklin Stahl. Published 04/08/2017
- ↑ Lenski, Richard. Convergence and Divergence in a Long-Term Experiment with Bacteria. The American Naturalist, Volume 190. August 2017. https://doi.org/10.1086/691209
Edited by [Claire Pruner], student of Joan Slonczewski for BIOL 116, 2024, Kenyon College.
