Saccharophagus degradans: Difference between revisions
(New page: Genus larsen From MicrobeWiki, the student-edited microbiology resource Jump to: navigation, search A Microbial Biorealm page on the genus Genus larsen Contents [hide] * 1 Classifica...) |
No edit summary |
||
| (43 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
Genus | {{Uncurated}} | ||
{{Biorealm Genus}} | |||
==Classification== | |||
===Higher order taxa=== | |||
Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae | |||
===Species=== | |||
Species | |||
''Saccharophagus degradans'' | |||
==Description and significance== | |||
Description and significance | [[Image:Mdeg_72.jpg|frame|right|''Saccharophagus degradans'' 2-40. From [http://genome.jgi-psf.org/finished_microbes/micde/micde.home.html DOE Joint Genome Institute.]]] | ||
''Saccharophagus degradans'' refers to a species of marine bacteria formally known as ''Microbulbifer degradans''. So far the only known strain is 2-40. It is a rod-shaped, aerobic, gram-negative, and motile γ-proteobacterium that was first isolated from decaying salt marsh cord grass ''Spartina alterniflora'' found in the Chesapeake Bay (2,3). ''S. degradans'' was isolated by pressing partially decomposed ''S. alterniflora'' into 1% peptone-half-strength-seawater agar plates. It was found that sea salt and carbohydrates were needed for growth of the bacteria. In addition, temperature range for growth was found to be 5 to 40°C with an optimum of 37°C while optimum pH was 7.5 (6). ''S. degradans'' is related to a group of marine bacteria that are responsible for the degradation of complex polysaccharides found in the ocean. This group includes the ''Microbulbifer'', ''Teredinibacter'', and ''Saccharophagus'' bacteria. Very little is known about how the almost annual 25 billion tons of CPs produced by organisms such as algae, land plants, crustaceans, bacteria and fungi are recycled back into usable carbon (1,4). Because of its relative importance to the marine carbon cycle, the US Department of Energy deemed it important to sequencing the genome of ''S. degradans'' 2-40 (1). ''S. degradans'' is unique in that it is able to degrade at least 10 different complex polysaccharides (CP) including agar, alginate, cellulose, chitin, β-glucan, laminarin, pectin, pullulan, starch, and xylan (2). What makes it even more amazing is that it is able to use all 10 CPs as its sole carbon and energy source. Although ''S. degradans'' was initially isolated in 1988, significant research was not done on it till 2005 when the genome was sequenced. | |||
==Genome structure== | |||
''Saccharophagus degradans'' has a circular chromosome that is 5,057,531 bp long. There are 4,008 protein coding genes out of a total of 4,067. The genome also includes 50 structural RNAs (5). What originally set ''S. degradans'' apart from the ''Microbulbifer'' and ''Teredinibacter'' groups was that it’s G+C content was 45.8% as compared to the 57-59% and 49-51% of ''Microbulbifer'' and ''Teredinibacter'' bacteria respectively (1). So far there have been identified 180 open reading frames (ORF) that code for carbohydrases. There are also around 112 ORFs that contain catalytic and/or carbohydrate-binding modules (CBM) with specificity for plant-derived polysaccharides (3). This is unique in that the number of ORFs is relatively large compared with other organisms with the same CP degrading functions. It is also worth noting that the size of the chromosome is relatively small for an organism with such a versatile polysaccharide degrading system. There are no known plasmids associated with ''Saccharophagus degradans'' that have been identified at this point. | |||
==Cell structure and metabolism== | |||
''Saccharophagus degradans'' is a gram-negative bacterium which means that it has a cell membrane in addition to a very thick cell wall on top of its membrane. The cell wall provides additional protection against hydrophobic substances like antibiotics. A structural characteristic that led to ''S. degradans'' being placed into its own distinct group was that the major fatty acid used was different compared to its relatives in the ''Microbulbifer'' and ''Teredinibacter'' groups. While iso-C<sub>15:0</sub> is used in ''Microbulbifer'' and ''Teredinibacter'' bacteria, ''S. degradans'' uses iso-C<sub>16:0</sub> (1). ''S. degradans'' mainly uses CPs as a source of metabolism. These include agar, alginate, cellulose, chitin, β-glucan, laminarin, pectin, pullulan, starch, and xylan (2). Although it can use all ten of these, the agar and cellulose pathways have been most studied. | |||
===Cellulose metabolism=== | |||
In cellulose degradation, 13 cellulose depolymerases are used accompanied by seven accessory enzymes which include two cellodextrinases, three cellobiases, a cellodextrin phosphorlyase, and a cellobiose phosphorylase (3). In one pathway, the cellulose are cleaved into shorter, random-length chains and cellodextrins. The shorter chains are further hydrolyzed by surface lipoproteins into cellodextrins. The cellodextrins are then cleaved into glucose and oligocellodextrins which are imported into the cytoplasm (3). Although there are other pathways that ''S. degradans'' uses to break down cellulose, it is not very well understood how all of the pathways work together in cellulose metabolism (3). | |||
===Agar metabolism=== | |||
Agar degradation is only partially understood in ''S. degradans''. What is known is that there are 5 identified agarases which are Aga50A, Aga50D, Aga86C, Aga86E, and Aga16B (2). All five of these are secreted and work together to break down agar using a β-agarase pathway. Agar polymers are first cleaved into an oligosaccharide derivative and then processed into a disaccharide product (2). An unusual feature was found Aga16B and Aga86E. Both of these showed the presence of multiple 6 carbohydrate binding modules (CBM6) with Aga16B having two homologs of CBM6 and Aga86E having three (2). CBMs are common in carbohydrases but are extremely rare in agarases. In fact, only two other agarases were found to have CBMs (2). | |||
==Ecology== | |||
''Saccharophagus degradans'' performs a vital role in the marine carbon cycle. It is part of an emerging group of bacteria that is responsible for the degradation of CPs produced by other organisms in the ocean. Since ''S. degradans'' has 10 distinct CP-degrading systems with more carbohydrases and accessory proteins than any marine bacterium studied so far (3), it is easily one of the most versatile CP degraders. This versatility helps keep the many different CPs from accumulating in the ocean. In the case of cellulose degradation, often a multitude of microorganisms are required. However, ''S. degradans'' can completely degrade cellulose by itself (3). This is important for places where ''S. degradans'' may be the only cellulose degrader. | |||
==Pathology== | |||
There are no known pathological characteristics of ''Saccharophagus degradans''. | |||
==Application to Biotechnology== | |||
The enzymes which allow ''S. degradans'' to break down 10 different CPs are constantly being isolated and studied. As the human population continues to grow, more pressure is being put on food production. As a result of the increased production, agricultural, aquacultural, and algalcultural wastes are starting to become serious problems. Cellulose, chitin, and agar are the major waste products. Using ''S. degradans'' as a powerful bioremediation tool may help curb the increase in waste products. Also, CPs can be hydrolyzed into usable feedstock. For many developing countries that have severe shortages of feedstock, research is being made into harnessing ''S. degradans''’ hydrolyzing power (4). | |||
==Current Research== | |||
===''S. degradans'' as a versatile complex polysaccharide degrader=== | |||
The analysis of the completed genome of ''Microbulbifer degradans'' 2-40 led to it being renamed to ''Saccharophagus degradans'' 2-40 and placed into its own genus ''Saccharophagus''. Research found that the G+C content was relatively lower at 45.8% as compared to its relatives in the ''Microbulbifer'' and ''Teredinibacter'' genus. The major fatty acid used by ''S. degradans'' was iso-C<sub>16:0</sub> while the other two genuses used iso-C<sub>15:0</sub>. ''S. degradans'' was tested along with other related species in the ''Microbulbifer'' genus and ''Teredinibacter turnerae'' to observe polysaccharide polymerization. Agar, alginate, cellulose, chitin, fucoidan, laminarin, pectin, pullulan, starch, and xylan were used in the experiment. The organisms were grown in sea salt liquid medium and then tested on each polysaccharide. ''S. degradans'' was the only one that showed depolymerization of all 10 polysaccharides (1). | |||
===Analyses of the Agarolytic system in ''S. degradans''=== | |||
A recent study was done to isolate the proteins and genes responsible for the agar degradation found in ''S. degradans''. Cells were grown in agar cultures. Mass spectrometry revealed three agarases from these cultures which were Aga16B, Aga86C, and Aga86E. Two more were revealed using deletion analysis. A total of 5 different agarases were isolated with Aga16B and Aga86E being shown to contain CBM6 domains which were unusual for agarases. The research revealed that the system consisted of secreted endolytic agarases and surface β-agarases (2). | |||
===Cellulase system of ''S. degradans''=== | |||
Current research into ''S. degradans'' CP degrading system has revealed that it can grow on cellulose alone without any help from other microorganisms. Cellulose degradation often requires a number of microorganisms working together to completely break it down. This prompted researchers to find out what was responsible for ''S. degradans''’ unique ability to grow on pure culture. The bacteria were grown on cellulose culture and then lysed to isolate the cellulose depolymerases. Mass spectrometry was then used to identify and match the enzymes to known ones. The researched resulted in the identification of 13 cellulose depolymerases that were complemented with seven accessory enzymes (3). | |||
==References== | |||
1. '''Ekborg, N. A., Gonzalez, J. M., Howard, M. B., Taylor, L. E., Hutcheson, S. W., and Weiner, R. M.''' "''Saccarophagus degradans'' gen. nov., sp. nov., a versatile marine degrader of complex polysaccharides". ''International Journal of Systematic and Evolutionary Microbiology''. 2005. Volume 55. p. 1545-1549. [http://ijs.sgmjournals.org/cgi/content/full/55/4/1545 http://ijs.sgmjournals.org/cgi/content/full/55/4/1545] | |||
2. '''Ekborg, N. A., Taylor, L. E., Longmire, A. G., Henrissat, B., Weiner, R. M., and Hutcheson, S. W. '''"Genomic and Proteomic Analyses of the Agarolytic System Expressed by ''Saccharophagus degradans'' 2-40". ''Applied and Environmental Microbiology''. May 2006. Volume 72. No. 5. p. 3396-3405. [http://aem.asm.org/cgi/content/full/72/5/3396?view=long&pmid=16672483 http://aem.asm.org/cgi/content/full/72/5/3396?view=long&pmid=16672483] | |||
3. '''Taylor, L. E., Henrissat, B., Coutinho, P. M., Ekborg, N. A., Hutcheson, S. W., and Weiner, R. M.''' "Complete Cellulase System in the Marine Bacterium ''Saccharophagus degradans'' Strain 2-40". ''Journal of Bacteriology''. June 2006. Volume 118. No. 11. p. 3849-3861. [http://jb.asm.org/cgi/content/full/188/11/3849?view=long&pmid=16707677 http://jb.asm.org/cgi/content/full/188/11/3849?view=long&pmid=16707677] | |||
4. '''JGI Saccharophagus degradans 2-40 Home'''. US Department of Energy Joint Genome Institute. 2005. [http://genome.jgi-psf.org/finished_microbes/micde/micde.home.html http://genome.jgi-psf.org/finished_microbes/micde/micde.home.html] | |||
5. '''''Saccharophagus degradans'''''. genus. NCBI reference: [http://www.ncbi.nlm.nih.gov/sites/entrez?Db=genome&Cmd=ShowDetailView&TermToSearch=19331 http://www.ncbi.nlm.nih.gov/sites/entrez?Db=genome&Cmd=ShowDetailView&TermToSearch=19331] | |||
6. '''Andrykovitch, G. and Marx, I.''' "Isolation of a New Polysaccharide-Digesting Bacterium from a Salt March". ''Applied and Environmental Microbiology''. April 1988. Volume 54. No. 4. p. 1061-1062. [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=16347602 http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=16347602] | |||
Edited by Cheng Liang Wu, student of [mailto:ralarsen@ucsd.edu Rachel Larsen] | |||
Edited by KLB | |||
Latest revision as of 03:33, 20 August 2010
A Microbial Biorealm page on the genus Saccharophagus degradans
Classification
Higher order taxa
Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae
Species
Saccharophagus degradans
Description and significance
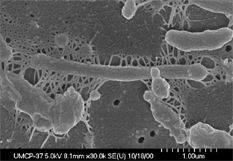
Saccharophagus degradans refers to a species of marine bacteria formally known as Microbulbifer degradans. So far the only known strain is 2-40. It is a rod-shaped, aerobic, gram-negative, and motile γ-proteobacterium that was first isolated from decaying salt marsh cord grass Spartina alterniflora found in the Chesapeake Bay (2,3). S. degradans was isolated by pressing partially decomposed S. alterniflora into 1% peptone-half-strength-seawater agar plates. It was found that sea salt and carbohydrates were needed for growth of the bacteria. In addition, temperature range for growth was found to be 5 to 40°C with an optimum of 37°C while optimum pH was 7.5 (6). S. degradans is related to a group of marine bacteria that are responsible for the degradation of complex polysaccharides found in the ocean. This group includes the Microbulbifer, Teredinibacter, and Saccharophagus bacteria. Very little is known about how the almost annual 25 billion tons of CPs produced by organisms such as algae, land plants, crustaceans, bacteria and fungi are recycled back into usable carbon (1,4). Because of its relative importance to the marine carbon cycle, the US Department of Energy deemed it important to sequencing the genome of S. degradans 2-40 (1). S. degradans is unique in that it is able to degrade at least 10 different complex polysaccharides (CP) including agar, alginate, cellulose, chitin, β-glucan, laminarin, pectin, pullulan, starch, and xylan (2). What makes it even more amazing is that it is able to use all 10 CPs as its sole carbon and energy source. Although S. degradans was initially isolated in 1988, significant research was not done on it till 2005 when the genome was sequenced.
Genome structure
Saccharophagus degradans has a circular chromosome that is 5,057,531 bp long. There are 4,008 protein coding genes out of a total of 4,067. The genome also includes 50 structural RNAs (5). What originally set S. degradans apart from the Microbulbifer and Teredinibacter groups was that it’s G+C content was 45.8% as compared to the 57-59% and 49-51% of Microbulbifer and Teredinibacter bacteria respectively (1). So far there have been identified 180 open reading frames (ORF) that code for carbohydrases. There are also around 112 ORFs that contain catalytic and/or carbohydrate-binding modules (CBM) with specificity for plant-derived polysaccharides (3). This is unique in that the number of ORFs is relatively large compared with other organisms with the same CP degrading functions. It is also worth noting that the size of the chromosome is relatively small for an organism with such a versatile polysaccharide degrading system. There are no known plasmids associated with Saccharophagus degradans that have been identified at this point.
Cell structure and metabolism
Saccharophagus degradans is a gram-negative bacterium which means that it has a cell membrane in addition to a very thick cell wall on top of its membrane. The cell wall provides additional protection against hydrophobic substances like antibiotics. A structural characteristic that led to S. degradans being placed into its own distinct group was that the major fatty acid used was different compared to its relatives in the Microbulbifer and Teredinibacter groups. While iso-C15:0 is used in Microbulbifer and Teredinibacter bacteria, S. degradans uses iso-C16:0 (1). S. degradans mainly uses CPs as a source of metabolism. These include agar, alginate, cellulose, chitin, β-glucan, laminarin, pectin, pullulan, starch, and xylan (2). Although it can use all ten of these, the agar and cellulose pathways have been most studied.
Cellulose metabolism
In cellulose degradation, 13 cellulose depolymerases are used accompanied by seven accessory enzymes which include two cellodextrinases, three cellobiases, a cellodextrin phosphorlyase, and a cellobiose phosphorylase (3). In one pathway, the cellulose are cleaved into shorter, random-length chains and cellodextrins. The shorter chains are further hydrolyzed by surface lipoproteins into cellodextrins. The cellodextrins are then cleaved into glucose and oligocellodextrins which are imported into the cytoplasm (3). Although there are other pathways that S. degradans uses to break down cellulose, it is not very well understood how all of the pathways work together in cellulose metabolism (3).
Agar metabolism
Agar degradation is only partially understood in S. degradans. What is known is that there are 5 identified agarases which are Aga50A, Aga50D, Aga86C, Aga86E, and Aga16B (2). All five of these are secreted and work together to break down agar using a β-agarase pathway. Agar polymers are first cleaved into an oligosaccharide derivative and then processed into a disaccharide product (2). An unusual feature was found Aga16B and Aga86E. Both of these showed the presence of multiple 6 carbohydrate binding modules (CBM6) with Aga16B having two homologs of CBM6 and Aga86E having three (2). CBMs are common in carbohydrases but are extremely rare in agarases. In fact, only two other agarases were found to have CBMs (2).
Ecology
Saccharophagus degradans performs a vital role in the marine carbon cycle. It is part of an emerging group of bacteria that is responsible for the degradation of CPs produced by other organisms in the ocean. Since S. degradans has 10 distinct CP-degrading systems with more carbohydrases and accessory proteins than any marine bacterium studied so far (3), it is easily one of the most versatile CP degraders. This versatility helps keep the many different CPs from accumulating in the ocean. In the case of cellulose degradation, often a multitude of microorganisms are required. However, S. degradans can completely degrade cellulose by itself (3). This is important for places where S. degradans may be the only cellulose degrader.
Pathology
There are no known pathological characteristics of Saccharophagus degradans.
Application to Biotechnology
The enzymes which allow S. degradans to break down 10 different CPs are constantly being isolated and studied. As the human population continues to grow, more pressure is being put on food production. As a result of the increased production, agricultural, aquacultural, and algalcultural wastes are starting to become serious problems. Cellulose, chitin, and agar are the major waste products. Using S. degradans as a powerful bioremediation tool may help curb the increase in waste products. Also, CPs can be hydrolyzed into usable feedstock. For many developing countries that have severe shortages of feedstock, research is being made into harnessing S. degradans’ hydrolyzing power (4).
Current Research
S. degradans as a versatile complex polysaccharide degrader
The analysis of the completed genome of Microbulbifer degradans 2-40 led to it being renamed to Saccharophagus degradans 2-40 and placed into its own genus Saccharophagus. Research found that the G+C content was relatively lower at 45.8% as compared to its relatives in the Microbulbifer and Teredinibacter genus. The major fatty acid used by S. degradans was iso-C16:0 while the other two genuses used iso-C15:0. S. degradans was tested along with other related species in the Microbulbifer genus and Teredinibacter turnerae to observe polysaccharide polymerization. Agar, alginate, cellulose, chitin, fucoidan, laminarin, pectin, pullulan, starch, and xylan were used in the experiment. The organisms were grown in sea salt liquid medium and then tested on each polysaccharide. S. degradans was the only one that showed depolymerization of all 10 polysaccharides (1).
Analyses of the Agarolytic system in S. degradans
A recent study was done to isolate the proteins and genes responsible for the agar degradation found in S. degradans. Cells were grown in agar cultures. Mass spectrometry revealed three agarases from these cultures which were Aga16B, Aga86C, and Aga86E. Two more were revealed using deletion analysis. A total of 5 different agarases were isolated with Aga16B and Aga86E being shown to contain CBM6 domains which were unusual for agarases. The research revealed that the system consisted of secreted endolytic agarases and surface β-agarases (2).
Cellulase system of S. degradans
Current research into S. degradans CP degrading system has revealed that it can grow on cellulose alone without any help from other microorganisms. Cellulose degradation often requires a number of microorganisms working together to completely break it down. This prompted researchers to find out what was responsible for S. degradans’ unique ability to grow on pure culture. The bacteria were grown on cellulose culture and then lysed to isolate the cellulose depolymerases. Mass spectrometry was then used to identify and match the enzymes to known ones. The researched resulted in the identification of 13 cellulose depolymerases that were complemented with seven accessory enzymes (3).
References
1. Ekborg, N. A., Gonzalez, J. M., Howard, M. B., Taylor, L. E., Hutcheson, S. W., and Weiner, R. M. "Saccarophagus degradans gen. nov., sp. nov., a versatile marine degrader of complex polysaccharides". International Journal of Systematic and Evolutionary Microbiology. 2005. Volume 55. p. 1545-1549. http://ijs.sgmjournals.org/cgi/content/full/55/4/1545
2. Ekborg, N. A., Taylor, L. E., Longmire, A. G., Henrissat, B., Weiner, R. M., and Hutcheson, S. W. "Genomic and Proteomic Analyses of the Agarolytic System Expressed by Saccharophagus degradans 2-40". Applied and Environmental Microbiology. May 2006. Volume 72. No. 5. p. 3396-3405. http://aem.asm.org/cgi/content/full/72/5/3396?view=long&pmid=16672483
3. Taylor, L. E., Henrissat, B., Coutinho, P. M., Ekborg, N. A., Hutcheson, S. W., and Weiner, R. M. "Complete Cellulase System in the Marine Bacterium Saccharophagus degradans Strain 2-40". Journal of Bacteriology. June 2006. Volume 118. No. 11. p. 3849-3861. http://jb.asm.org/cgi/content/full/188/11/3849?view=long&pmid=16707677
4. JGI Saccharophagus degradans 2-40 Home. US Department of Energy Joint Genome Institute. 2005. http://genome.jgi-psf.org/finished_microbes/micde/micde.home.html
5. Saccharophagus degradans. genus. NCBI reference: http://www.ncbi.nlm.nih.gov/sites/entrez?Db=genome&Cmd=ShowDetailView&TermToSearch=19331
6. Andrykovitch, G. and Marx, I. "Isolation of a New Polysaccharide-Digesting Bacterium from a Salt March". Applied and Environmental Microbiology. April 1988. Volume 54. No. 4. p. 1061-1062. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=16347602
Edited by Cheng Liang Wu, student of Rachel Larsen
Edited by KLB
