Sodalis glossinidius: Difference between revisions
(New page: {{Biorealm Genus}} ==Classification== ===Higher order taxa=== Domain; Phylum; Class; Order; family [Others may be used. Use [http://www.ncbi.nlm.nih.gov/Taxonomy/ NCBI] link to find] ...) |
No edit summary |
||
| (136 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | |||
{{Biorealm Genus}} | {{Biorealm Genus}} | ||
| Line 5: | Line 6: | ||
===Higher order taxa=== | ===Higher order taxa=== | ||
Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae (1) | |||
===Species=== | ===Species=== | ||
''Sodalis glossinidius'' (1) | |||
''' | |||
'' | ==Description and significance== | ||
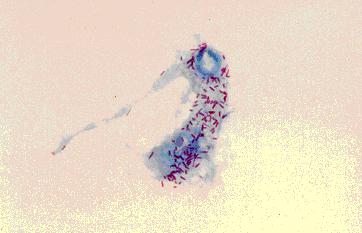
[[Image:Tsetse_haemocyte_and_S_glossinidius.JPG|frame|right|Symbiotic lifestyle of ''S. glossinidius'' (purple) in a haemocyte of the tsetse fly. Image courtesy of Prof. Sue Welburn, University of Edinburgh.]] | |||
''Sodalis glossinidius'' is a Gram-negative, nonspore-forming, rod-shaped, filamentous bacteria. It has flagella expressed during immature host developmental stages, though it is non-motile(10). It is one of three endosymbionts for the tsetse fly (''Glossina'' spp.), which are all maternally transmitted to progeny(6,7). The other two being ''Wigglesworthia glossinidium'' and a strain of ''Wolbachia''(5). This secondary endosymbiont has a mutualistic relationship with the tsetse fly and resides inter- and intracellularly in the midgut, fat body and haemolymph of the insect(7). ''S. glossinidius'' was the first true insect endosymbiont to be isolated and cultured, achieved in 1999 by Colin Dale and Ian Maudlin at the University of Glasgow. This was performed using a mosquito (''Aedes albopictus'') feeder cell culture system and a sample from the haemolymph of ''Glossinia morsitans morsitans''(7). The type strain M1T was isolated(7). ''S. glossinidius'' can be cultured intracellularly in ''Aedes albopictus'' or axenically in semidefined solid medium containing already enzimatically digested proteins as a nitrogen source. This microaerophilic bacterium grows optimally with 5% oxygen and 95% carbon dioxide at 25°C. Colony morphology is uniform and off-white with defined edges.(7). | |||
The tsetse fly is a vector for ''Trypanosoma brucei gambiense'' and ''Trypanosoma brucei rhodesiense''(9). These parasitic protozoa cause African trypanosomiasis (commonly known as African Sleeping Sickness), an epidemic fatal disease plaguing sub-Saharan Africa(9). ''S. glossinidius'' is important enough to have its genome sequenced because this it is a key player involved in the vector-parasite relationship, possibly enabling the creation of tsetse flies that are incapable of transimtting trypanosomes and thus controlling the spread of African sleeping sickness (See Ecology for more details)(6). The genome of ''S. glossinidius'' also provides an example of an organism that has undergone degenerative adaptations in order to adopt a mutualistic relationship with its host (See Genome structure for more details)(3). | |||
==Genome structure== | ==Genome structure== | ||
''Sodalis glossinidius'' has one circular chromosome approximately 4 Mb(10). This micro-organism has a relatively inactive biochemical profile (2,432 protein coding sequences, indicating 51% reduced coding capacity) compared to other organisms in the family Enterobacteriaceae(3,7,10). This is likely due to its evolutionary path away from free-existence. The genome reduction and function loss undergone by ''S. glossinidius'' is less than other intracellular pathogens and obligate symbionts, shown by its ability to be cultured ''in vitro'', but its genome size is still smaller than closely related free-living enterics(2). This further suggests that it is in the process of becoming more dependent on the tsetse host cell. Though most organisms of the Family Enterobacteriaceae are able to produce catalase, ''S. glossinidius'' is not; this thought to be why this micro-organism requires a microaerophilic environment(7). Genes dealing with transcription, translation, regulation, and nucleic acid and amino acid biosynthetic pathways were retained. On the other hand, 972 pseudogenes (mostly caused by single point mutations) are homologs of known proteins for defense or for transporting and metabolizing carbohydrates and inorganic ions, reflecting the mutualistic relationship with the host(2,10). | |||
The genome of ''S. glossinidius'' has an A+T content of only 45%, whereas endosymbiont genomes are typically AT-rich. The A+T content of intracellular pathogen ''R. prowazekii'' is 71% and 75% for the mutualist ''Buchnera''. The AT-rich genomes of intracellular pathogens and obligates may be due to the loss of function of DNA repair and recombination functions, suggesting that ''S. glossinidius'' has retained these genes(2). The 16S rRNA of ''S. glossinidius'' shares high sequence identity with other secondary endosymbionts of insects. In addition, it was found to be almost identical within the species when isolated from different ''Glossina'' spp. This suggests recent acquisition by the tsetse as an enteric and possible horizontal symbiont transfer(7). | |||
Extrachromosomal DNA consists of three plasmids, pSG1 (82 kb), pSG2 (27 kb), and pSG4 (11 kb) and a prophage-like element, pSOG3 (52 kb)(4,12). pSOG3 retains complete ORFs required for virion assembly, with nothing intact in the chromosomal sequence(12). pSG1, pSG2, and pSG4 all have putative RepA proteins. Plasmid pSG1 codes for the production and transport of siderophores which aid in the acquisition of iron(4). This CDS is located between transposase open reading frames (ORFs) indicating horizontal gene transfer(4). Plasmid pSG1 also contains ORFs with homology to effectors, toxins, hemolysins, and protease fitness factors(4). There were several regions with homology to conjugative transfer pilus genes suggesting that conjugation has been important for ''S. glossinidius''(10). Tra1 region on pSG1 had ORFs similar to transfer regions of plasmids. On the same plasmid is Tra2, but both regions were disrupted by mutation. On plasmid pSG2 was a region with homology to pilus ORFs(10). | |||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
''S. glossinidius'' is Gram-negative and rod-shaped(7). The lipopolysaccharide in this bacteria seems to be expressed without the O-antigen. This reduces the micro-organism's resistance to host immunity, another indicator of the mutualistic relationship between host and symbiont(5). Extrachromosomal DNA encodes siderophores for the acquisition of iron(4). Though ''S. glossinidius'' is non-motile, there are 90 flagellar-related coding sequences on the chromosome. A gene cluster for a complete flagellar apparatus including motility proteins (MotA and MotB) is present(10). | |||
There are also three phylogenetically distinct type-III secretion systems (TTSSs) referred to as ''Sodalis'' symbiosis regions (SSRs) designated SSR-1, SSR-2, and SSR-3(10). When assembled on the surface of the bacterial cell, the multicomponent TTSS functions as a molecular syringe so the bacteria can inject effector proteins into the host cell and then to facilitate its own entry(3). While core components of the needle structure were conserved, regions coding for some effectors or regulators have been modified or eliminated(10). SSR-1 is similar to the pathogenicity island ''ysa'' from ''Yersinia enterocolitica'', which is involved in entry into the host cell. SSR-2 is similar to the ''Salmonella'' pathogenicity island SPI-1 and Mxi-Spa from ''Shigella flexneri'', which are involved in post-infection processes once the bacteria has already entered. SSR-3 is similar to SPI-2, which is found in both ''Salmonella'' spp. and ''Yersinia pestis'', and is crucial for survival and virulence(5,10). The TTSSs and flagellar components are expressed during early larval development. They are thought to allow ''S. glossinidius'' to enter host cells and ensure the establishment and maintenance of symbiosis in intra-uterine progeny(10). | |||
''S. glossinidius'' has retained functional pathways for glycolysis, gluconeogenesis, the TCA cycle and the pentose phosphate pathway of free-living bacteria(10). Many energy metabolism and carbon compound assimilation genes are missing in the genome. This may indicate an adaptation to the low concentration and lack of diversity of carbohydrates available due to vertebrate blood being the only nutrient source of the tsetse host(2,5). The pathways to synthesize all amino acids except alanine are present(10). Most genes involved in energy production and conversion via anaerobic respiration have been lost. Which is expected in the constant microaerophilic environment of the tsetse midgut and haemolymph(10). In its iron-rich environment, iron transport across the cytoplasmic membrane is achieved through active transport using an ABC transporter(10). It also has an intact FeoA ferrous iron importer. Furthermore, plasmid pSG1 has a system of siderophores to transport and store iron(4). | |||
The principle carbon source during growth is N-acetyl-d-glucosamine (Glc''N''Ac), which it produces from the breakdown of chitin(6). Chitin (polymerized N-acetyl-d-glucosamine) is present in the exoskeleton and gut peritrophic membrane of the tsetse fly and ''S. glossinidius'' catabolizes it by producing the chitinase, b-N-acetylglucosaminidase(7). | |||
==Ecology== | ==Ecology== | ||
[[Image:Fly.jpg|frame|right|Female tsetse fly. Image courtesy of United States Department of Agriculture.]] | |||
''S. glossinidius'' is an endosymbiont of the tsetse fly with a mutualistic relationship(''T. b. gambiense'')(7). Symbionts to blood-feeding insects are thought to supplement the restricted diet or blood by providing cofactors and vitamin metabolites. Genes for biotin and lipoic acid, molybdenum cofactor, thiamine, riboflavin, and folic acid were present in the micro-organism's genome(2). ''S. glossinidius'' is known to increase trypanosome infection rate for tsetse(6). (See Pathology for more details) | |||
When plated ''in vitro'', the beginning of the streak where the inoculum was heavy, produced many large colonies that merged. Toward the end of the streak, where innoculum was thinner, colonies were less abundant and smaller(7). Like other microaerophilic bacteria, ''S. glossinidius'' has population-dependent growth where growth rate increases with total respiratory capacity(7). | |||
==Pathology== | ==Pathology== | ||
''Sodalis glossinidius'' is a mutualist micro-organism to tsetse and its chromosomally encoded TTSSs, hemolysin, lipases, and adhesions are thought to fulfill functions different from those reported in pathogenic microbes(10). Despite this mutualistic relationship, it has been shown that ''S. glossinidius'' does increase susceptibility to trypanosome infection for the tsetse host by hindering the tsetse immune system(6). When tsetse were treated with streptozotocin (2-deoxy-2-(3-methyl-3-nitrosoureido)--glucopyranoside), a bacteriocidal analogue of Glc''N''Ac (primary carbon source of ''S. glossinidius''), the fecundity and pupal emergence rates were largely unaffected(6). This antibiotic does not affect the other symbionts of ''T. b. gambiense''. Glc''N''Ac is an effective inhibitor of tsetse midgut lectin, which has a role in non-self recognition and combatting tsetse fly infection(11). These lectin inhibitory sugars accumulate in tsetse puparia through the enzymatic activity of chitinases secreted by ''S. glossinigius'', thus promoting increased trypanosome infection rates in tsetse(6,10). | |||
==Application to Biotechnology== | ==Application to Biotechnology== | ||
''S. glossinidius'' has lost much of its biochemical profile(7). Genetic material for compounds and enzymes have drifted into pseudogene status due to its specialized environment, the tsetse fly(3.) There are no known biotechnological benefits to society. | |||
==Current Research== | ==Current Research== | ||
Some of the recent research on ''Sodalis glossinidius'': | |||
'''A Possible Heterodimeric Prophage-Like Element in the Genome of the Insect Endosymbiont Sodalis glossinidius'''(12) | |||
Though bacterial symbionts rarely contain bacteriophages due to their lack of contact outside the host. ''Sodalis glossinidius'' contains an extrachomosomal element pSOG3 with phage-related sequences. A culture from ''Glossina morsitans morsitans'' was isolated and pSOG3 was shotgun sequenced. It was found to be 52 kb with 50 putative CDSs and five pseudogenes. This extrachromosomal element is thought to have formed from the integration of two distinct ancestral phage genomes. This heterodimeric element was derived from extant phages HK620 (a relative of P22) and epsilon15. Degeneration occured when ''S. glossinidius'' moved toward symbiosis with tsetse. pSOG3 is thought to replicate extrachromosomally. There are two sequences hypothesized to be involved in replication. The putative protein product of an ORF is similar to RepA in ''Erwinia amylovora''. Another ORF is similar to the plasmid partitioning protein, ParA, of ''Corynebacterium glutamicum''. Phage virions have been identified sporadically in S. glossinidius and it is hypothesized that pSOG3 is the prophage responsible for those virions. | |||
'''Interspecific Transfer of Bacterial Endosymbionts between Tsetse Fly Species: Infection Establishment and Effect on Host Fitness'''(8) | |||
This research is exploring how to use transgenic technologies in the hopes of diminishing vectorial capacity of the tsetse fly via genetic manipulation. To control trypanosomiasis, paratransgenesis has already been developed where a foreign protein is expressed in ''S. glossinidius'' that are toxic to trypanosomes. This endosymbiont is ideal for paratransgenesis because of it's vertical transmission to progeny. Research is aimed to determine the level of biological integration of ''S. glossinidius'' and tsetse. ''G. fuscipes fuscipes'' and ''G. morsitans morsitans'' flies were cleared of their native ''Sodalis'' symbionts and then transinfected with ''Sodalis'' from a different tsetse fly species in order to streamline the development of paratransgenesis. Quantitative PCR (QT-PCR) analysis was used to compare ''Sodalis'' symbiont densities in the control and transinfected tsetse flies. It was found that endogenous wildtype bacteria did not have a competitive advantage over recombinant native and nonnative bacteria. The results also went against the previous hypothesis that there is a positive correlation between high Sodalis densities and trypanosome infection rates. Based on symbiont densities, this study found that the degree of susceptibility to infection may not correlate solely with the Sodalis density. | |||
'''A novel phospholipase from ''Trypanosoma brucei'''''(13) | |||
Phospholipase A1 (PLA1) (EC 3.1.1.32) specifically hydrolyses the sn-1 acyl esters from phospholipids producing free fatty acids and lysophospholipids. Its activity has been recorded in a wide range of organisms, but very few PLA1 has been cloned and characterized. The phospholipase A1 in ''Trypanosoma brucei'' (TbPLA1) is unique because it is evolutionarily related to bacterial secreted PLA1. It was most likely acquired by horizontal gene transfer from ''Sodalis glossinidius'', sharing 21.5% primary sequence. It was acquired once a parasitic lifestyle with the tsetse host was established. There are no eukaryotic homologues of TbPLA1. The bacterial PLA1 possess a signal sequence for targeted secretion, but is absent in TbPLA1. This suggested the intracellular localization of PLA1 in ''T. brucei''. | |||
==References== | ==References== | ||
[ | 1. "''Sodalis glossinidius''". <u>NCBI Taxonomy Browser</u>. 26 August 2007. [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=63612&lvl=3&lin=f&keep=1&srchmode=1&unlock] | ||
2. Akman, L., Rio, R., Beard, C., and Aksoy, S. “Genome Size Determination and Coding Capacity of ''Sodalis glossinidius'', an Enteric Symbiont of Tsetse Flies, as Revealed by Hybridization to Escherichia coli Gene Arrays.” ''Journal of Bacteriology''. 2001. Volume 183.15 p. 4517-4525.[http://jb.asm.org/cgi/content/full/183/15/4517] | |||
3. Dale, C., Jones, T., and Pontes, M. "Degenerative Evolution and Functional Diversification of Type-III Secretion Systems in the Insect Endosymbiont ''Sodalis glossinidius''." ''Molecular Biology and Evolution''. 2005. Volume 22.3 p. 758-766. [http://mbe.oxfordjournals.org/cgi/content/full/22/3/758] | |||
4. Darby, A., Lagnel, J., Matthew, C., Bourtzis, K., Maudlin, I., and Welburn, S. "Extrachromosomal DNA of the Symbiont ''Sodalis glossinidius''." ''Journal of Bacteriology''. 2005. Volume 187.14 p. 5003-5007. [http://jb.asm.org/cgi/content/full/187/14/5003?view=long&pmid=15995217] | |||
5. Thomson, N., Crossman, L., and Bentley, S. "Bacterial home from home." ''Nature Reviews Microbiology''. 2006. Volume 4 p. 168-170. [http://www.nature.com/nrmicro/journal/v4/n3/full/nrmicro1367.html] | |||
6. Dale, C., and Welburn, S. "The endosymbionts of tsetse flies: manipulating host–parasite interactions." ''International Journal for Parasitology''. 2001. Volume 31.5-6 p. 627-630. [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6T7F-42WX3BM-11&_user=4429&_coverDate=05%2F01%2F2001&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000059602&_version=1&_urlVersion=0&_userid=4429&md5=32e639fc02894be83034e1718707992f#bib2] | |||
7. Dale, C., and Maudlin, I. "Sodalis gen. nov. and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly Glossina morsitans morsitans." ''International Journal of Systematic Bacteriology''. 1999. Volume 49 p. 267–275. [http://ijs.sgmjournals.org/cgi/reprint/49/1/267] | |||
8. Weiss, B., Mouchotte, R., Rio, R., Wu, Y., Wu, Z., Heddi, A., and Aksoy, S. "Interspecific Transfer of Bacterial Endosymbionts between Tsetse Fly Species: Infection Establishment and Effect on Host Fitness." ''Applied and Environmental Microbiology''. Nov 2006. Volume 72.11 p. 7013-7021. [http://aem.asm.org/cgi/content/full/72/11/7013?view=long&pmid=16950907] | |||
9. Maudlin, I. "African trypanosomiasis." ''Annals of Tropical Medicine & Parasitology''. 2006. Volume 11.8 p. 679–701. [http://docserver.ingentaconnect.com/deliver/connect/maney/00034983/v100n8/s4.pdf?expires=1188478216&id=39237070&titleid=6120&accname=University+of+California+San+Diego&checksum=D499BF4D0DFD13CEBCF2BABD912CE539] | |||
10. Toh, H., Weiss, B., Perkin, S., Yamashita, A., Oshima, K., Hattori, M., and Aksoy, S. "Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of '''Sodalis glossinidius'' in the tsetse host." ''Genome Research''. 2006. Volume 16 p. 149-156. [http://www.genome.org/cgi/content/full/16/2/149] | |||
11. Basseri, H. "Role of Lectins in Interaction Between Parasites and the Important Insect Vectors." ''Iranian J. Publ. Health''. 2002. Volume 31.1-2 p. 69-74. [http://diglib.tums.ac.ir/pub/magmng/pdf/326.pdf] | |||
12. Clark, A., Pontes, M., Jones, T., and Dale, C. "A Possible Heterodimeric Prophage-Like Element in the Genome of the Insect Endosymbiont ''Sodalis glossinidius''." ''J Bacteriol.'' 2007. Volume 189.7 p. 2949–2951. [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=17209029] | |||
13. Richmond, G., and Smith, T. "A novel phospholipase from ''Trypanosoma brucei''." ''Molecular Microbiology''. 2007. Volume 63.4 p. 1078-1095.[http://www.blackwell-synergy.com/doi/full/10.1111/j.1365-2958.2006.05582.x] | |||
Edited by Janet Melnyk, student of [mailto:ralarsen@ucsd.edu Rachel Larsen] | |||
Edited by | Edited by KLB | ||
Latest revision as of 03:34, 20 August 2010
A Microbial Biorealm page on the genus Sodalis glossinidius
Classification
Higher order taxa
Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae (1)
Species
Sodalis glossinidius (1)
Description and significance
Sodalis glossinidius is a Gram-negative, nonspore-forming, rod-shaped, filamentous bacteria. It has flagella expressed during immature host developmental stages, though it is non-motile(10). It is one of three endosymbionts for the tsetse fly (Glossina spp.), which are all maternally transmitted to progeny(6,7). The other two being Wigglesworthia glossinidium and a strain of Wolbachia(5). This secondary endosymbiont has a mutualistic relationship with the tsetse fly and resides inter- and intracellularly in the midgut, fat body and haemolymph of the insect(7). S. glossinidius was the first true insect endosymbiont to be isolated and cultured, achieved in 1999 by Colin Dale and Ian Maudlin at the University of Glasgow. This was performed using a mosquito (Aedes albopictus) feeder cell culture system and a sample from the haemolymph of Glossinia morsitans morsitans(7). The type strain M1T was isolated(7). S. glossinidius can be cultured intracellularly in Aedes albopictus or axenically in semidefined solid medium containing already enzimatically digested proteins as a nitrogen source. This microaerophilic bacterium grows optimally with 5% oxygen and 95% carbon dioxide at 25°C. Colony morphology is uniform and off-white with defined edges.(7).
The tsetse fly is a vector for Trypanosoma brucei gambiense and Trypanosoma brucei rhodesiense(9). These parasitic protozoa cause African trypanosomiasis (commonly known as African Sleeping Sickness), an epidemic fatal disease plaguing sub-Saharan Africa(9). S. glossinidius is important enough to have its genome sequenced because this it is a key player involved in the vector-parasite relationship, possibly enabling the creation of tsetse flies that are incapable of transimtting trypanosomes and thus controlling the spread of African sleeping sickness (See Ecology for more details)(6). The genome of S. glossinidius also provides an example of an organism that has undergone degenerative adaptations in order to adopt a mutualistic relationship with its host (See Genome structure for more details)(3).
Genome structure
Sodalis glossinidius has one circular chromosome approximately 4 Mb(10). This micro-organism has a relatively inactive biochemical profile (2,432 protein coding sequences, indicating 51% reduced coding capacity) compared to other organisms in the family Enterobacteriaceae(3,7,10). This is likely due to its evolutionary path away from free-existence. The genome reduction and function loss undergone by S. glossinidius is less than other intracellular pathogens and obligate symbionts, shown by its ability to be cultured in vitro, but its genome size is still smaller than closely related free-living enterics(2). This further suggests that it is in the process of becoming more dependent on the tsetse host cell. Though most organisms of the Family Enterobacteriaceae are able to produce catalase, S. glossinidius is not; this thought to be why this micro-organism requires a microaerophilic environment(7). Genes dealing with transcription, translation, regulation, and nucleic acid and amino acid biosynthetic pathways were retained. On the other hand, 972 pseudogenes (mostly caused by single point mutations) are homologs of known proteins for defense or for transporting and metabolizing carbohydrates and inorganic ions, reflecting the mutualistic relationship with the host(2,10).
The genome of S. glossinidius has an A+T content of only 45%, whereas endosymbiont genomes are typically AT-rich. The A+T content of intracellular pathogen R. prowazekii is 71% and 75% for the mutualist Buchnera. The AT-rich genomes of intracellular pathogens and obligates may be due to the loss of function of DNA repair and recombination functions, suggesting that S. glossinidius has retained these genes(2). The 16S rRNA of S. glossinidius shares high sequence identity with other secondary endosymbionts of insects. In addition, it was found to be almost identical within the species when isolated from different Glossina spp. This suggests recent acquisition by the tsetse as an enteric and possible horizontal symbiont transfer(7).
Extrachromosomal DNA consists of three plasmids, pSG1 (82 kb), pSG2 (27 kb), and pSG4 (11 kb) and a prophage-like element, pSOG3 (52 kb)(4,12). pSOG3 retains complete ORFs required for virion assembly, with nothing intact in the chromosomal sequence(12). pSG1, pSG2, and pSG4 all have putative RepA proteins. Plasmid pSG1 codes for the production and transport of siderophores which aid in the acquisition of iron(4). This CDS is located between transposase open reading frames (ORFs) indicating horizontal gene transfer(4). Plasmid pSG1 also contains ORFs with homology to effectors, toxins, hemolysins, and protease fitness factors(4). There were several regions with homology to conjugative transfer pilus genes suggesting that conjugation has been important for S. glossinidius(10). Tra1 region on pSG1 had ORFs similar to transfer regions of plasmids. On the same plasmid is Tra2, but both regions were disrupted by mutation. On plasmid pSG2 was a region with homology to pilus ORFs(10).
Cell structure and metabolism
S. glossinidius is Gram-negative and rod-shaped(7). The lipopolysaccharide in this bacteria seems to be expressed without the O-antigen. This reduces the micro-organism's resistance to host immunity, another indicator of the mutualistic relationship between host and symbiont(5). Extrachromosomal DNA encodes siderophores for the acquisition of iron(4). Though S. glossinidius is non-motile, there are 90 flagellar-related coding sequences on the chromosome. A gene cluster for a complete flagellar apparatus including motility proteins (MotA and MotB) is present(10).
There are also three phylogenetically distinct type-III secretion systems (TTSSs) referred to as Sodalis symbiosis regions (SSRs) designated SSR-1, SSR-2, and SSR-3(10). When assembled on the surface of the bacterial cell, the multicomponent TTSS functions as a molecular syringe so the bacteria can inject effector proteins into the host cell and then to facilitate its own entry(3). While core components of the needle structure were conserved, regions coding for some effectors or regulators have been modified or eliminated(10). SSR-1 is similar to the pathogenicity island ysa from Yersinia enterocolitica, which is involved in entry into the host cell. SSR-2 is similar to the Salmonella pathogenicity island SPI-1 and Mxi-Spa from Shigella flexneri, which are involved in post-infection processes once the bacteria has already entered. SSR-3 is similar to SPI-2, which is found in both Salmonella spp. and Yersinia pestis, and is crucial for survival and virulence(5,10). The TTSSs and flagellar components are expressed during early larval development. They are thought to allow S. glossinidius to enter host cells and ensure the establishment and maintenance of symbiosis in intra-uterine progeny(10).
S. glossinidius has retained functional pathways for glycolysis, gluconeogenesis, the TCA cycle and the pentose phosphate pathway of free-living bacteria(10). Many energy metabolism and carbon compound assimilation genes are missing in the genome. This may indicate an adaptation to the low concentration and lack of diversity of carbohydrates available due to vertebrate blood being the only nutrient source of the tsetse host(2,5). The pathways to synthesize all amino acids except alanine are present(10). Most genes involved in energy production and conversion via anaerobic respiration have been lost. Which is expected in the constant microaerophilic environment of the tsetse midgut and haemolymph(10). In its iron-rich environment, iron transport across the cytoplasmic membrane is achieved through active transport using an ABC transporter(10). It also has an intact FeoA ferrous iron importer. Furthermore, plasmid pSG1 has a system of siderophores to transport and store iron(4).
The principle carbon source during growth is N-acetyl-d-glucosamine (GlcNAc), which it produces from the breakdown of chitin(6). Chitin (polymerized N-acetyl-d-glucosamine) is present in the exoskeleton and gut peritrophic membrane of the tsetse fly and S. glossinidius catabolizes it by producing the chitinase, b-N-acetylglucosaminidase(7).
Ecology
S. glossinidius is an endosymbiont of the tsetse fly with a mutualistic relationship(T. b. gambiense)(7). Symbionts to blood-feeding insects are thought to supplement the restricted diet or blood by providing cofactors and vitamin metabolites. Genes for biotin and lipoic acid, molybdenum cofactor, thiamine, riboflavin, and folic acid were present in the micro-organism's genome(2). S. glossinidius is known to increase trypanosome infection rate for tsetse(6). (See Pathology for more details)
When plated in vitro, the beginning of the streak where the inoculum was heavy, produced many large colonies that merged. Toward the end of the streak, where innoculum was thinner, colonies were less abundant and smaller(7). Like other microaerophilic bacteria, S. glossinidius has population-dependent growth where growth rate increases with total respiratory capacity(7).
Pathology
Sodalis glossinidius is a mutualist micro-organism to tsetse and its chromosomally encoded TTSSs, hemolysin, lipases, and adhesions are thought to fulfill functions different from those reported in pathogenic microbes(10). Despite this mutualistic relationship, it has been shown that S. glossinidius does increase susceptibility to trypanosome infection for the tsetse host by hindering the tsetse immune system(6). When tsetse were treated with streptozotocin (2-deoxy-2-(3-methyl-3-nitrosoureido)--glucopyranoside), a bacteriocidal analogue of GlcNAc (primary carbon source of S. glossinidius), the fecundity and pupal emergence rates were largely unaffected(6). This antibiotic does not affect the other symbionts of T. b. gambiense. GlcNAc is an effective inhibitor of tsetse midgut lectin, which has a role in non-self recognition and combatting tsetse fly infection(11). These lectin inhibitory sugars accumulate in tsetse puparia through the enzymatic activity of chitinases secreted by S. glossinigius, thus promoting increased trypanosome infection rates in tsetse(6,10).
Application to Biotechnology
S. glossinidius has lost much of its biochemical profile(7). Genetic material for compounds and enzymes have drifted into pseudogene status due to its specialized environment, the tsetse fly(3.) There are no known biotechnological benefits to society.
Current Research
Some of the recent research on Sodalis glossinidius:
A Possible Heterodimeric Prophage-Like Element in the Genome of the Insect Endosymbiont Sodalis glossinidius(12) Though bacterial symbionts rarely contain bacteriophages due to their lack of contact outside the host. Sodalis glossinidius contains an extrachomosomal element pSOG3 with phage-related sequences. A culture from Glossina morsitans morsitans was isolated and pSOG3 was shotgun sequenced. It was found to be 52 kb with 50 putative CDSs and five pseudogenes. This extrachromosomal element is thought to have formed from the integration of two distinct ancestral phage genomes. This heterodimeric element was derived from extant phages HK620 (a relative of P22) and epsilon15. Degeneration occured when S. glossinidius moved toward symbiosis with tsetse. pSOG3 is thought to replicate extrachromosomally. There are two sequences hypothesized to be involved in replication. The putative protein product of an ORF is similar to RepA in Erwinia amylovora. Another ORF is similar to the plasmid partitioning protein, ParA, of Corynebacterium glutamicum. Phage virions have been identified sporadically in S. glossinidius and it is hypothesized that pSOG3 is the prophage responsible for those virions.
Interspecific Transfer of Bacterial Endosymbionts between Tsetse Fly Species: Infection Establishment and Effect on Host Fitness(8) This research is exploring how to use transgenic technologies in the hopes of diminishing vectorial capacity of the tsetse fly via genetic manipulation. To control trypanosomiasis, paratransgenesis has already been developed where a foreign protein is expressed in S. glossinidius that are toxic to trypanosomes. This endosymbiont is ideal for paratransgenesis because of it's vertical transmission to progeny. Research is aimed to determine the level of biological integration of S. glossinidius and tsetse. G. fuscipes fuscipes and G. morsitans morsitans flies were cleared of their native Sodalis symbionts and then transinfected with Sodalis from a different tsetse fly species in order to streamline the development of paratransgenesis. Quantitative PCR (QT-PCR) analysis was used to compare Sodalis symbiont densities in the control and transinfected tsetse flies. It was found that endogenous wildtype bacteria did not have a competitive advantage over recombinant native and nonnative bacteria. The results also went against the previous hypothesis that there is a positive correlation between high Sodalis densities and trypanosome infection rates. Based on symbiont densities, this study found that the degree of susceptibility to infection may not correlate solely with the Sodalis density.
A novel phospholipase from Trypanosoma brucei(13) Phospholipase A1 (PLA1) (EC 3.1.1.32) specifically hydrolyses the sn-1 acyl esters from phospholipids producing free fatty acids and lysophospholipids. Its activity has been recorded in a wide range of organisms, but very few PLA1 has been cloned and characterized. The phospholipase A1 in Trypanosoma brucei (TbPLA1) is unique because it is evolutionarily related to bacterial secreted PLA1. It was most likely acquired by horizontal gene transfer from Sodalis glossinidius, sharing 21.5% primary sequence. It was acquired once a parasitic lifestyle with the tsetse host was established. There are no eukaryotic homologues of TbPLA1. The bacterial PLA1 possess a signal sequence for targeted secretion, but is absent in TbPLA1. This suggested the intracellular localization of PLA1 in T. brucei.
References
1. "Sodalis glossinidius". NCBI Taxonomy Browser. 26 August 2007. [1]
2. Akman, L., Rio, R., Beard, C., and Aksoy, S. “Genome Size Determination and Coding Capacity of Sodalis glossinidius, an Enteric Symbiont of Tsetse Flies, as Revealed by Hybridization to Escherichia coli Gene Arrays.” Journal of Bacteriology. 2001. Volume 183.15 p. 4517-4525.[2]
3. Dale, C., Jones, T., and Pontes, M. "Degenerative Evolution and Functional Diversification of Type-III Secretion Systems in the Insect Endosymbiont Sodalis glossinidius." Molecular Biology and Evolution. 2005. Volume 22.3 p. 758-766. [3]
4. Darby, A., Lagnel, J., Matthew, C., Bourtzis, K., Maudlin, I., and Welburn, S. "Extrachromosomal DNA of the Symbiont Sodalis glossinidius." Journal of Bacteriology. 2005. Volume 187.14 p. 5003-5007. [4]
5. Thomson, N., Crossman, L., and Bentley, S. "Bacterial home from home." Nature Reviews Microbiology. 2006. Volume 4 p. 168-170. [5]
6. Dale, C., and Welburn, S. "The endosymbionts of tsetse flies: manipulating host–parasite interactions." International Journal for Parasitology. 2001. Volume 31.5-6 p. 627-630. [6]
7. Dale, C., and Maudlin, I. "Sodalis gen. nov. and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly Glossina morsitans morsitans." International Journal of Systematic Bacteriology. 1999. Volume 49 p. 267–275. [7]
8. Weiss, B., Mouchotte, R., Rio, R., Wu, Y., Wu, Z., Heddi, A., and Aksoy, S. "Interspecific Transfer of Bacterial Endosymbionts between Tsetse Fly Species: Infection Establishment and Effect on Host Fitness." Applied and Environmental Microbiology. Nov 2006. Volume 72.11 p. 7013-7021. [8]
9. Maudlin, I. "African trypanosomiasis." Annals of Tropical Medicine & Parasitology. 2006. Volume 11.8 p. 679–701. [9]
10. Toh, H., Weiss, B., Perkin, S., Yamashita, A., Oshima, K., Hattori, M., and Aksoy, S. "Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of 'Sodalis glossinidius in the tsetse host." Genome Research. 2006. Volume 16 p. 149-156. [10]
11. Basseri, H. "Role of Lectins in Interaction Between Parasites and the Important Insect Vectors." Iranian J. Publ. Health. 2002. Volume 31.1-2 p. 69-74. [11]
12. Clark, A., Pontes, M., Jones, T., and Dale, C. "A Possible Heterodimeric Prophage-Like Element in the Genome of the Insect Endosymbiont Sodalis glossinidius." J Bacteriol. 2007. Volume 189.7 p. 2949–2951. [12]
13. Richmond, G., and Smith, T. "A novel phospholipase from Trypanosoma brucei." Molecular Microbiology. 2007. Volume 63.4 p. 1078-1095.[13]
Edited by Janet Melnyk, student of Rachel Larsen
Edited by KLB