Citrobacter freundii: Difference between revisions
No edit summary |
|||
| (60 intermediate revisions by 6 users not shown) | |||
| Line 1: | Line 1: | ||
{{Curated}} | |||
{{Biorealm Genus}} | {{Biorealm Genus}} | ||
| Line 23: | Line 24: | ||
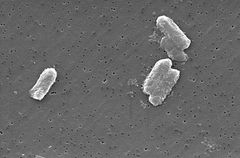
The Citrobacter species, including Citrobacter freundii, are aerobic gram-negative bacilli. Citrobacter freundii are long rod-shaped bacteria typically 1-5 μm in length [1]. Most C. freundii cells are surrounded by many flagella used to move about, but a few are non-motile. Its habitat includes the environment (soil, water, sewage), food, and the intestinal tracts of animals and humans [1]. It belongs to the family of Enterobacteriaceae. | The Citrobacter species, including Citrobacter freundii, are aerobic gram-negative bacilli. Citrobacter freundii are long rod-shaped bacteria typically 1-5 μm in length [1]. Most C. freundii cells are surrounded by many flagella used to move about, but a few are non-motile. Its habitat includes the environment (soil, water, sewage), food, and the intestinal tracts of animals and humans [1]. It belongs to the family of Enterobacteriaceae. | ||
As an opportunistic | As an opportunistic pathogen, C. freundii is responsible for a number of significant opportunistic infections. It is known to be the cause of a variety of nosocomial infections of the respiratory tract, urinary tract, blood and several other normally sterile sites in patients [2]. C. freundii represents approximately 29% of all opportunistic infections [2]. Therefore, one of the chief reasons many different strains and plasmids of the C. freundii genome are being sequenced is in order to find antibiotics that can fight these opportunistic infections. | ||
Surprisingly, this infectious microbe in humans plays a positive role in the environment. C. freundii is responsible for reducing nitrate to nitrite in the environment [3]. This crucial conversion is an important stage in the nitrogen cycle. And recycling nitrogen is very essential because the earth's atmosphere is about 85% nitrogen [3]. Therefore, due to its important contribution to the environment is another motivation for sequencing the genome of C. freundii. | Surprisingly, this infectious microbe in humans plays a positive role in the environment. C. freundii is responsible for reducing nitrate to nitrite in the environment [3]. This crucial conversion is an important stage in the nitrogen cycle. And recycling nitrogen is very essential because the earth's atmosphere is about 85% nitrogen [3]. Therefore, due to its important contribution to the environment is another motivation for sequencing the genome of C. freundii. | ||
| Line 31: | Line 32: | ||
==Genome structure== | ==Genome structure== | ||
No information about the complete genome of C. freundii is available online, although some individual strains and plasmids of the microbe have been sequenced. The most prominent one is the plasmid pCTX-M#3 because it is the largest plasmid and encodes a large amount of proteins. Its sequence was completed on January 6, 2005. It is a circular DNA plasmid and it is 89,468 nucleotide base pairs long. The length of the plasmid is 0.089468 (Mbp). It is composed of 51.0 % GC content, and encodes 105 proteins [4]. | |||
Another important feature of the C. freundii genome is that it is the only microbe in the Enterobacteriaceae family that contains a plasmid which encodes L-methionine γ-lyase (MGL). The nucleotide sequence of the plasmid contains a 3000 bp long EcoRI insert [5]. The fragment also contains two open reading frames. The first frame consists of 1,194 nucleotides, and the second, 1,296 nucleotides.The first frame, known as the megL gene, encodes a protein of 398 amino acid residues that has sequence homology with MGLs from different sources. The second frame encodes a protein with sequence homology with proteins belonging to the family of permeases [5]. | Another important feature of the C. freundii genome is that it is the only microbe in the Enterobacteriaceae family that contains a plasmid which encodes L-methionine γ-lyase (MGL). The nucleotide sequence of the plasmid contains a 3000 bp long EcoRI insert [5]. The fragment also contains two open reading frames. The first frame consists of 1,194 nucleotides, and the second, 1,296 nucleotides.The first frame, known as the megL gene, encodes a protein of 398 amino acid residues that has sequence homology with MGLs from different sources. The second frame encodes a protein with sequence homology with proteins belonging to the family of permeases [5]. | ||
The C. freundii OS60 AmpC β-lactamase gene has also been sequenced and it is composed of 1197 nucleotides. It encodes a 380 amino acid long precursor and contains a 19 residue signal peptide in the 5’ end [6]. This gene encodes a mature protein that has a molecular mass of 39 781 Daltons. | The C. freundii OS60 AmpC β-lactamase gene has also been sequenced and it is composed of 1197 nucleotides. It encodes a 380 amino acid long precursor and contains a 19 residue signal peptide in the 5’ end [6]. This gene encodes a mature protein that has a molecular mass of 39 781 Daltons. | ||
The amino acid positions in these precursor are suprisinlgy identical to residues in the E. coli K12 chromosomal AmpC β-lactamases | The amino acid positions in these precursor are suprisinlgy identical to residues in the E. coli K12 chromosomal AmpC β-lactamases [6]. | ||
Another important strain in the genome of C. freunii is GN346, which is a clinical isolate recovered in 1965. This strain produces the enzyme cephalosporinase, which has the ability to hydrolyzes and inactivate the anibitoics cephalosporins and cephamycins [7]. The structural and promoter regions of the cephalosporinase gene are 1408 nucleotides long. The amino acid sequence of the mature enzyme, is composed of 361 amino acids with a molecular mass of 39,878 Da [7]. | Another important strain in the genome of C. freunii is GN346, which is a clinical isolate recovered in 1965. This strain produces the enzyme cephalosporinase, which has the ability to hydrolyzes and inactivate the anibitoics cephalosporins and cephamycins [7]. The structural and promoter regions of the cephalosporinase gene are 1408 nucleotides long. The amino acid sequence of the mature enzyme, is composed of 361 amino acids with a molecular mass of 39,878 Da [7]. | ||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
The cell structure of C. | The cell structure of C. freundi is long and rod-shaped usually 1-5 μm in length. The outside of the cell contains many flagella used for motality [1]. Since C. freundii is gram-negative bacteria, it contains two membranes (inner and outer).The periplasmic space lies in between the two membranes. The outer membrane does not contain an energy source; but it does contain many porins embedded within that help the organism acquire important ions [1]. Unlike gram-positive bacteria, C. freundii cells do not contain a thick cell wall made up of peptidoglycan. | ||
For metabolism, C. freundii has an amazing ability to grow on glycerol as the sole carbon and energy source. In this process, glycerol is fermented by a dismutation process. This process requires two pathways [8]. In the first pathway, glycerol is dehydrogenated by a NAD1-linked glycerol dehydrogenase to dihydroxyacetone. The dihydroxyacetone is then phosphorylated and funneled to glycolysis by dihydroxyacetone kinase [8]. In the second pathway, glycerol is dehydrated by the coenzyme B12-dependent glycerol dehydratase to form 3-hydroxypropionaldehyde [8]. This product is reduced to the major fermentation product 1,3-propanediol by the NADH-linked 1,3-propanediol dehydrogenase, which regenerates NAD1. The dha regulon encodes the four essential enzymes of these two pathways. Amazingly, the expression of the dha regulon is only induced when glycerol is present [8]. | |||
Cells of C. freundii are also able to | Cells of C. freundii are also able to metabolize lactose or citrate as a carbon source [8]. | ||
==Ecology== | ==Ecology== | ||
Citrobacter freundii | Citrobacter freundii are commonly found in the environment, mainly in soil, water, and sewages. They are an indicator of potential contamination of water. They are also found on different organs of diseased animals, including mammals, birds, reptiles, and amphibians [1]. They are not known to interact with other organims. | ||
In the environment, C. freundii can convert nitrate or the ammonium ion (which is a nitrogen atom combined with four hydrogen atoms) to nitrite; this reaction occurs in the environment as well as within the digestive tract of humans and other animals. After it converts | In the environment, C. freundii can convert nitrate or the ammonium ion (which is a nitrogen atom combined with four hydrogen atoms) to nitrite; this reaction occurs in the environment as well as within the digestive tract of humans and other animals [3]. After it converts nitrate to nitrite in the environment, the nitrite is converted to nitrogen, and this final step completes the nitrogen cycle in the earth's atmosphere, which is made up of 85% nitrogen [3]. This organism's ecological role not only includes its important role in the nitrogen cycle, because it can also accumulate uranium (which is the basic material for nuclear technology) by building phosphate complexes [3]. | ||
Citrobacter freundii has also been investigated for biodegradation of tannic acid used in tannerys. | Citrobacter freundii has also been investigated for biodegradation of tannic acid used in tannerys [3]. | ||
==Pathology== | ==Pathology== | ||
As an opportunistic pathogen, Citrobacter freundii is often the cause of significant opportunistic infections, meaning that it does not generally cause disease in healthy | As an opportunistic pathogen, Citrobacter freundii is often the cause of significant opportunistic infections, meaning that it does not generally cause disease in healthy human hosts. They only affect patients with a weak immune system, signifying that they need an "opportunity" to infect the person [2]. Therefore, in patients with a suppressed immune system, Citrobacter species are known to cause a wide variety of nosocomial infections of the respiratory tract, urinary tract, and the blood [2]. Hepatic, biliary and pancreatic disease are also common diseases that are caused by C. freundii. The biliary tract is the most common site of infection by the C. freundii bacilli [9]. | ||
Citrobacter species cause a wide variety of nosocomial infections of the respiratory tract, | |||
urinary tract, blood | |||
One fatal disease that C. freundii has | One fatal disease that C. freundii has been associated with is neonatal meningitis. Neonatal meningitis is the inflammation of the meninges (the system of membranes which surround the CNS) due to bacterial invasion [10]. The mortality rate of Citrobacter meningitis is unacceptably high, with death rates of patients ranging from 25 to 50 %. Moreover, serious neurological problems still persist in 75% of survivors. In this disease, Citrobacter freundii is able to penetrate the blood-brain barrier that consists of the choroid plexus epithelium and the brain capillary endothelium [10]. | ||
The diseases | Tests performed by Badger et. al in the article “Citrobacter freundii Invades and Replicates in Human Brain Microvascular Endothelial Cells”suggest that bacterial proliferation of C. freundii takes place at the intracellular level, which had been contrary to the general scientific thought. The findings indicate that C. freundii traverses vacuoles, replicates and is released into the basolateral side of the human brain microvascular endothelial cells (HBMEC) in order to cross the blood-brain barrier. Further analysis may potentially allow for therapeutic strategies to treat infections. There is still no therapeutic treatment available [22]. | ||
Certain diseases studied in trout and cyprinids are also caused by C. freundii. C. freundii causes abnormal inflammatory changes in the intestine of trout and inflammatory and necrotic changes in the internal organs of cyprinids. The illness was discovered by means of artificial infection with a pure culture of C. freundii. This discovery established C. freundii as a cause of fish disease [11]. | |||
In a case study by the Journal of Medical Microbiology, a patient developed peritonitis and tunnel infection due to Citrobacter freundii which is uncommon. The patient was on continuous ambulatory peritoneal dialysis. Usually the causing agents are gram-positive micro-organisms, particularly Staphylococcus aureus and Staphyloccus edpidermis. Also there are no known reports of tunnel infection due to C. freundii. Initial antibiotic therapy did not work and the infection continued to persist until the catheter was removed. This is clinically significant because Citrobacter Freundii show different antibiotic susceptibility which is why initial therapy was not successful. The patient did not respond to treatment until the catheter was removed showing Citrobacter freundii are opportunistic pathogens that affect hospitalized and immunocompromised patients [18]. | |||
==Application to Biotechnology== | ==Application to Biotechnology== | ||
In the Biotech | In the Biotech industry, Citrobacter freundii produces many important enzymes. | ||
The first enzymes it produces is phosphatase. | The first enzymes it produces is phosphatase. Phosphatase activity of C. freundii has been postulated to be involved in lead accumulation, which can have play an important role in the Biotech industry. The phosphatase activity of C. freundii has been also discovered to have resistance to some diagnostic reagents [16]. | ||
The | The purification and characterization of bacterial selenocysteine beta-lyase, an enzyme which specifically catalyzes the cleavage of L-selenocysteine to L-alanine, has been purified from Citrobacter freundii. The enzyme is monomeric with a molecular weight of ca. 64,000 and contains 1 mol of pyridoxal 5'-phosphate as a cofactor per mol of enzyme. The enzyme also catalyzes the alpha, beta elimination of beta-chloro-L-alanine to form NH3, pyruvate [15]. | ||
C. Freundii strains also carry a plasmid that encodes class 1 AmpC cephalosporinase. These enzymes can hydrolzye inactivate new cephamycins and cephalosporins. | C. Freundii strains also carry a plasmid that encodes class 1 AmpC cephalosporinase. These enzymes can hydrolzye inactivate new cephamycins and cephalosporins [17]. | ||
==Current Research== | ==Current Research== | ||
A small scale research concerning certain strains of C. freundii was done recently at the University of Tennessee, Knoxville. The importance of certain tetracycline and streptomycin resistance genes and class 1 integrons in C. freundii isolated from dairy farm soil and nondairy soils | A small scale research concerning certain strains of C. freundii was done recently at the University of Tennessee, Knoxville. The importance of certain tetracycline and streptomycin resistance genes and class 1 integrons in C. freundii isolated from dairy farm soil and nondairy soils were evaluated. One strain of C. freundii extracted from dairy farm soils carried class 1 integrons with different inserted gene cassettes. Results of this small study suggested that the presence of multiple resistance genes and class 1 integrons in C. freundii in dairy farm soil may act as a reservoir of antimicrobial resistance genes and could play a role in the dissemination of these antimicrobial resistance genes to other commensal and indigenous microbial communities in soil. However, additional longer-term studies conducted in more locations are needed to support this hypothesis [12]. | ||
A second research concering C. freundii was done in order to devise a polymerase chain reaction (PCR) method that simultaneously uses three pairs of specific primers to detect genes of certain microbes (including C. freundii). The method included designing three primer pairs which were: SPVC-1 and SPVC-2, INVA-1 and INVA-2; and VIAB-1 and VIAB-2. PCR was performed using these three primers to identify 14 clinically important bacterial organisms. The following strains were quickly identified using the PCR: (1) C. freundii; (2) S. Typhi; and S. Paratyphi C; (3) S. Dublin (virulence antigen-positive); and (4) Salmonella serovars that harbor an spv-type virulence plasmid. Although this PCR method is new, with the advance of technology in the future this method can allow the identification of C. freundii in mammals immediately so that appropriate antibiotic treatment can be initiated without delay [13]. | |||
A third study concering C.freundii was done at the University of Barcelona, Spain. The mechanisms of resistance to fluoroquinolones in two Citrobacter freundii strains were studied. Both strains were isolated from the same patient. This study allowed partial characterisation of the acrA and acrB genes of this microorganism. Expression of genes in both strains was analysed using DNA microarrays for Escherichia coli. Nucleotide similarity between the partially sequenced acrA and acrB genes of C. freundii and E. coli was 80.7% and 85%, respectively. The acrA and acrB genes of C. freundii are similar to those in E. coli and their overexpression may play an important role in modulating the final minimum inhibitory concentration of fluoroquinolones [14]. | |||
A | A fourth study concerning C. freundii was done in Taiwan. A team of researchers isolated a diabetic patient that developed necrotizing fasciitis which was caused by C. freundii from an injury incited by a marine animal. Necrotizing fasciitis is an infection within the deeper layer of the skin and subcutaneous tissues. When treating the patient they took a sample from the fluid in the wound and found C. freundii. After three days of starting the antibiotic treatments with cefotaxmine and cefepmine, there was accumulation of subcutaneous abscesses. After 6, 10, 14, and 21 days of tending to the patient, some antibiotics were giving the patient some relief, but no long term recovery was reached. Cefotoaxime, cefepime, ciprofloxamin are among the antibiotics that were no match for C. freundii. The patient fully recovered after 42 days of ertapenem treatment. The researchers had isolated two colonies of C. freundii within 5 days of each other. As the isolates were grown they became immensely resistant to cefotaxime and cefepime. The reason why entrapenem worked against C. freundii is because is active against AmpC producing Enterobacteriaceae [21]. | ||
==Antibiotic resistance== | |||
Citrobacter species are a common cause of nosocomial infections associated with patients that are undergoing prolonged hospital treatments. C. freundii has recently been reported to express resistance to broad-spectrum antibiotincs including piperacillin, piperacillintazobactam, vancomycin and cephalosporins. Isolation of ceftriaxone-resistant Citrobacter freundii (CRCF) has been associated with the overprescribed broad spectrum antibiotics. The emerging new CRCF strains could suggest induction or depression of resistance genes as well as elimination of competing organisms. CRCF has been mostly isolated from patients with significant comorbidities including AIDS, peripheral vascular disease, and cerebrovascular disease. The usage of fluoroquinolone has also been reported to have no affect against the isolation of CRCF [19]. | |||
Citrobacter freundii is also known to contain in its chromosome a gene coding for cephalosporinase. This enzyme hydrolyses −CO−NH− bond in the lactam ring of cephalosporins and cephamycis thus rendering the bacteria resistant to this type of antibiotics. However when exposed to new third generation cephems and cerbapenems, clinically isolated C.freundii showed sensitivity to those substances. | |||
A small outbreak of C.freundii resistant to third generation cephems has been observed in the surgical ward of Nagoya University Hospital in patients that underwent surgical procedures. The C.freundii was isolated from patient's bile, wound gauze, feces, pus, and ascites. It was suggested that these new strains of C. freundii contained a plasmid encoding AmpC celphalosporinase but upon failure to transfer the cephems resistance from C. freundii to E.coli it was concluded that the enzyme must be encoded in the chromosome of C. freundii. Since C. freundi is associated with nosocomial infections caution to this new strains is recommended [20]. | |||
==References== | ==References== | ||
1. Wang JT,Chang SC, Chen YC, Luh KT. “Comparison of antimicrobial susceptibility of Citrobacter freundii isolates in two different time periods.” The Journal of Microbiology, Immunology and Infection. 2000 Dec; 33(4): 258-62 | 1. Wang JT,Chang SC, Chen YC, Luh KT. “Comparison of antimicrobial susceptibility of Citrobacter freundii isolates in two different time periods.” The Journal of Microbiology, Immunology and Infection. 2000 Dec; 33(4): 258-62. | ||
2. Whalen JG, Mully TW, Enlgish JC 3rd. “Spontaneous Citrobacter freundii infection in an immunocompetent patient.” Archives of dermatology. 2007 Jan; 143(1): 124-5 | 2. Whalen JG, Mully TW, Enlgish JC 3rd. “Spontaneous Citrobacter freundii infection in an immunocompetent patient.” Archives of dermatology. 2007 Jan; 143(1): 124-5. | ||
3. Puchenkova SG. “Enterobacteria in areas of water along the Crimean Coast.” Mikrobiolohichnyĭ zhurnal. 1996 Mar-Apr; 58(2): 3-7 | 3. Puchenkova SG. “Enterobacteria in areas of water along the Crimean Coast.” Mikrobiolohichnyĭ zhurnal. 1996 Mar-Apr; 58(2): 3-7. | ||
4.http://www.ncbi.nlm.nih.gov/sites/entrez?db=genomeprj&cmd=Retrieve&dopt=Overview&list_uids=13123 | 4.http://www.ncbi.nlm.nih.gov/sites/entrez?db=genomeprj&cmd=Retrieve&dopt=Overview&list_uids=13123 | ||
| Line 97: | Line 111: | ||
7. Tsukamoto K, Tachibana K, Yamazaki N, Ishii Y, Ujiie K, Nishida N, Sawai T. “Role of lysine-67 in the active site of class C beta-lactamase from Citrobacter freundii GN346.” European Journal of Biochemistry/FEBS. 1990 Feb 22; 188(1): 15-22. | 7. Tsukamoto K, Tachibana K, Yamazaki N, Ishii Y, Ujiie K, Nishida N, Sawai T. “Role of lysine-67 in the active site of class C beta-lactamase from Citrobacter freundii GN346.” European Journal of Biochemistry/FEBS. 1990 Feb 22; 188(1): 15-22. | ||
8. Keevil CW, Hough JS, Cole JA. “Prototrophic growth of Citrobacter freundii and the biochemical basis for its apparent growth requirements in aerated media.” Journal of General Microbiology. 1997 Jan; 98(1): 273-6. | |||
9. Marco Sánchez F, Turabian Fernández JL, Durán Pérez-Navarro A. “Fatal Citrobacter Freundii Bronchopneumonia Acquired in the Community in an Uncompromised Patient.” Revista Clínica Española. 1985 Apr; 176(6): 320 | |||
10. Julie L. Badger, Monique F. Stins, and Kwang Sik Kim. “Citrobacter freundii Invades and Replicates in Human Brain Microvascular Endothelial Cells.” Hinyokika kiyo. Acta Urologica Japonica. 1985 Jul; 31(7): 1159-70. | |||
11. Drelichman V, Band JD. “Bacteremias due to Citrobacter diversus and Citrobacter freundii. Incidence, risk factors, and clinical outcome.” Archives of Internal Medicine. 1985 Oct; 145(10): 1808-10. | |||
12. Srinivasan V, Nam HM, Sawant AA, Headrick SI, Nguyen LT, Oliver SP. “Distribution of Tetracycline and Streptomycin Resistance Genes and Class 1 Integrons in Enterobacteriaceae Isolated from Dairy and Nondairy Farm Soil.” | |||
Microbial Ecology. 2007 Aug 15. | |||
13. Sánchez-Céspedes J, Vila J. “Partial characterisation of the acrAB locus in two Citrobacter freundii clinical isolates.” International Journal of Antimicrobial Agents. 2007 Sep; 30(3): 259-63. | |||
14. Ciğerci IH, Korcan SE, Konuk M, Oztürk S. “Comparison of ALAD activities of Citrobacter and Pseudomonas strains and their usage as biomarker for Pb contamination.” Environmental Monitoring and Assessment. 2007 May 22. | |||
15. Chocat P, Esaki N, Tanizawa K, Nakamura K, Tanaka H, Soda K. “Purification and characterization of selenocysteine beta-lyase from Citrobacter freundii.” Journal of Bacteriology. 1985 Aug; 163(2): 669-76. | |||
16. Hillel S Levinson, Inga Mahler. “Phosphatase activity and lead resistance in Citrobacter freundii and Staphylococcus aureus.” FEMS Microbiology Letters. 1998 Apr; 161(1): 135-138. | |||
17. Murakami K, Yoshida T. “Covalent binding of moxalactam to cephalosporinase of Citrobacter freundii.” Antimicrobial Agents and Chemotherapy. 1985 May; 27(5): 727-32. | |||
18. Dervisoglu, E., Yegenaga, I., Yumuk, Z. “Citrobacter Freundii peritonitis and tunnel infection in a patient on continuous ambulatory peritoneal dialysis.” Journal of Medical Microbiology, 2008. Volume 57. p. 125-127. | |||
19. Kim PW, Harris AD, Roghmann MC, Morris JG Jr, Strinivasan A, Perencevich EN. "Epidemiological risk factors for isolation of ceftriaxone-resistant versus -susceptible citrobacter freundii in hospitalized patients" Antimicrob Agents Chemother, 2003 Sep;47(9): p.2882-2887. | |||
20. Nada T, Baba H, Kawamura K, Ohkura T, Torii K, Ohta M. "A small outbreak of third generation cephem-resistant Citrobacter freundii infection on a surgical ward" Jpn J Infect Dis. 2004 Aug;57(4): p.181-182. | |||
21. Chuang, Y., Tseng, S.,Teng, L., Ho, Y., and Hsueh, P. "Emergence of cefotaxime resistance in Citrobactor freundii causing necrotizing fasciitis and osteomyelitis" Journal of Infection, 2006. Volume 53. p. e161-e163. | |||
22.Badger, J., Stins, M., and Sik Kim, K. “Citrobacter freundii Invades and Replicates in Human Brain Microvascular Endothelial Cells” Infection and Immunity, 1999. Volume67. P. 4208-4215. | |||
Edited by Sumaira Akbarzada, student of [mailto:ralarsen@ucsd.edu Rachel Larsen] | Edited by Sumaira Akbarzada, student of [mailto:ralarsen@ucsd.edu Rachel Larsen] | ||
Edited by Greg Vargas and Darren Zhen ,students of [mailto:mglogow@luc.edu M Glogowski]at [http://www.luc.edu/biology/index.html Loyola University] | |||
Edited by Gergana Grigorova and Michal Olszewski, students of [mailto:mglogow@luc.edu M Glogowski]at [http://www.luc.edu/biology/index.html Loyola University] | |||
Edited by Alex Kula and Joanna Aiken, students of [mailto:mglogow@luc.edu M Glogowski]at [http://www.luc.edu/biology/index.html Loyola University] | |||
<!-- Do not edit or remove this line-->[[Category:Pages edited by students of Mary Glogowski at Loyola University]] | |||
Latest revision as of 19:08, 22 April 2011
A Microbial Biorealm page on the genus Citrobacter freundii
Classification
Higher order taxa
Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae; Citrobacter
Species
|
NCBI: Taxonomy |
Citrobacter Freundii
Description and significance
The Citrobacter species, including Citrobacter freundii, are aerobic gram-negative bacilli. Citrobacter freundii are long rod-shaped bacteria typically 1-5 μm in length [1]. Most C. freundii cells are surrounded by many flagella used to move about, but a few are non-motile. Its habitat includes the environment (soil, water, sewage), food, and the intestinal tracts of animals and humans [1]. It belongs to the family of Enterobacteriaceae.
As an opportunistic pathogen, C. freundii is responsible for a number of significant opportunistic infections. It is known to be the cause of a variety of nosocomial infections of the respiratory tract, urinary tract, blood and several other normally sterile sites in patients [2]. C. freundii represents approximately 29% of all opportunistic infections [2]. Therefore, one of the chief reasons many different strains and plasmids of the C. freundii genome are being sequenced is in order to find antibiotics that can fight these opportunistic infections.
Surprisingly, this infectious microbe in humans plays a positive role in the environment. C. freundii is responsible for reducing nitrate to nitrite in the environment [3]. This crucial conversion is an important stage in the nitrogen cycle. And recycling nitrogen is very essential because the earth's atmosphere is about 85% nitrogen [3]. Therefore, due to its important contribution to the environment is another motivation for sequencing the genome of C. freundii.
The Citrobacter genus was discovered in 1932 by Werkman and Gillen. Cultures of C. freundii were isolated and identified in the same year from soil extracts [1].
Genome structure
No information about the complete genome of C. freundii is available online, although some individual strains and plasmids of the microbe have been sequenced. The most prominent one is the plasmid pCTX-M#3 because it is the largest plasmid and encodes a large amount of proteins. Its sequence was completed on January 6, 2005. It is a circular DNA plasmid and it is 89,468 nucleotide base pairs long. The length of the plasmid is 0.089468 (Mbp). It is composed of 51.0 % GC content, and encodes 105 proteins [4].
Another important feature of the C. freundii genome is that it is the only microbe in the Enterobacteriaceae family that contains a plasmid which encodes L-methionine γ-lyase (MGL). The nucleotide sequence of the plasmid contains a 3000 bp long EcoRI insert [5]. The fragment also contains two open reading frames. The first frame consists of 1,194 nucleotides, and the second, 1,296 nucleotides.The first frame, known as the megL gene, encodes a protein of 398 amino acid residues that has sequence homology with MGLs from different sources. The second frame encodes a protein with sequence homology with proteins belonging to the family of permeases [5].
The C. freundii OS60 AmpC β-lactamase gene has also been sequenced and it is composed of 1197 nucleotides. It encodes a 380 amino acid long precursor and contains a 19 residue signal peptide in the 5’ end [6]. This gene encodes a mature protein that has a molecular mass of 39 781 Daltons. The amino acid positions in these precursor are suprisinlgy identical to residues in the E. coli K12 chromosomal AmpC β-lactamases [6].
Another important strain in the genome of C. freunii is GN346, which is a clinical isolate recovered in 1965. This strain produces the enzyme cephalosporinase, which has the ability to hydrolyzes and inactivate the anibitoics cephalosporins and cephamycins [7]. The structural and promoter regions of the cephalosporinase gene are 1408 nucleotides long. The amino acid sequence of the mature enzyme, is composed of 361 amino acids with a molecular mass of 39,878 Da [7].
Cell structure and metabolism
The cell structure of C. freundi is long and rod-shaped usually 1-5 μm in length. The outside of the cell contains many flagella used for motality [1]. Since C. freundii is gram-negative bacteria, it contains two membranes (inner and outer).The periplasmic space lies in between the two membranes. The outer membrane does not contain an energy source; but it does contain many porins embedded within that help the organism acquire important ions [1]. Unlike gram-positive bacteria, C. freundii cells do not contain a thick cell wall made up of peptidoglycan.
For metabolism, C. freundii has an amazing ability to grow on glycerol as the sole carbon and energy source. In this process, glycerol is fermented by a dismutation process. This process requires two pathways [8]. In the first pathway, glycerol is dehydrogenated by a NAD1-linked glycerol dehydrogenase to dihydroxyacetone. The dihydroxyacetone is then phosphorylated and funneled to glycolysis by dihydroxyacetone kinase [8]. In the second pathway, glycerol is dehydrated by the coenzyme B12-dependent glycerol dehydratase to form 3-hydroxypropionaldehyde [8]. This product is reduced to the major fermentation product 1,3-propanediol by the NADH-linked 1,3-propanediol dehydrogenase, which regenerates NAD1. The dha regulon encodes the four essential enzymes of these two pathways. Amazingly, the expression of the dha regulon is only induced when glycerol is present [8].
Cells of C. freundii are also able to metabolize lactose or citrate as a carbon source [8].
Ecology
Citrobacter freundii are commonly found in the environment, mainly in soil, water, and sewages. They are an indicator of potential contamination of water. They are also found on different organs of diseased animals, including mammals, birds, reptiles, and amphibians [1]. They are not known to interact with other organims.
In the environment, C. freundii can convert nitrate or the ammonium ion (which is a nitrogen atom combined with four hydrogen atoms) to nitrite; this reaction occurs in the environment as well as within the digestive tract of humans and other animals [3]. After it converts nitrate to nitrite in the environment, the nitrite is converted to nitrogen, and this final step completes the nitrogen cycle in the earth's atmosphere, which is made up of 85% nitrogen [3]. This organism's ecological role not only includes its important role in the nitrogen cycle, because it can also accumulate uranium (which is the basic material for nuclear technology) by building phosphate complexes [3].
Citrobacter freundii has also been investigated for biodegradation of tannic acid used in tannerys [3].
Pathology
As an opportunistic pathogen, Citrobacter freundii is often the cause of significant opportunistic infections, meaning that it does not generally cause disease in healthy human hosts. They only affect patients with a weak immune system, signifying that they need an "opportunity" to infect the person [2]. Therefore, in patients with a suppressed immune system, Citrobacter species are known to cause a wide variety of nosocomial infections of the respiratory tract, urinary tract, and the blood [2]. Hepatic, biliary and pancreatic disease are also common diseases that are caused by C. freundii. The biliary tract is the most common site of infection by the C. freundii bacilli [9].
One fatal disease that C. freundii has been associated with is neonatal meningitis. Neonatal meningitis is the inflammation of the meninges (the system of membranes which surround the CNS) due to bacterial invasion [10]. The mortality rate of Citrobacter meningitis is unacceptably high, with death rates of patients ranging from 25 to 50 %. Moreover, serious neurological problems still persist in 75% of survivors. In this disease, Citrobacter freundii is able to penetrate the blood-brain barrier that consists of the choroid plexus epithelium and the brain capillary endothelium [10].
Tests performed by Badger et. al in the article “Citrobacter freundii Invades and Replicates in Human Brain Microvascular Endothelial Cells”suggest that bacterial proliferation of C. freundii takes place at the intracellular level, which had been contrary to the general scientific thought. The findings indicate that C. freundii traverses vacuoles, replicates and is released into the basolateral side of the human brain microvascular endothelial cells (HBMEC) in order to cross the blood-brain barrier. Further analysis may potentially allow for therapeutic strategies to treat infections. There is still no therapeutic treatment available [22].
Certain diseases studied in trout and cyprinids are also caused by C. freundii. C. freundii causes abnormal inflammatory changes in the intestine of trout and inflammatory and necrotic changes in the internal organs of cyprinids. The illness was discovered by means of artificial infection with a pure culture of C. freundii. This discovery established C. freundii as a cause of fish disease [11].
In a case study by the Journal of Medical Microbiology, a patient developed peritonitis and tunnel infection due to Citrobacter freundii which is uncommon. The patient was on continuous ambulatory peritoneal dialysis. Usually the causing agents are gram-positive micro-organisms, particularly Staphylococcus aureus and Staphyloccus edpidermis. Also there are no known reports of tunnel infection due to C. freundii. Initial antibiotic therapy did not work and the infection continued to persist until the catheter was removed. This is clinically significant because Citrobacter Freundii show different antibiotic susceptibility which is why initial therapy was not successful. The patient did not respond to treatment until the catheter was removed showing Citrobacter freundii are opportunistic pathogens that affect hospitalized and immunocompromised patients [18].
Application to Biotechnology
In the Biotech industry, Citrobacter freundii produces many important enzymes. The first enzymes it produces is phosphatase. Phosphatase activity of C. freundii has been postulated to be involved in lead accumulation, which can have play an important role in the Biotech industry. The phosphatase activity of C. freundii has been also discovered to have resistance to some diagnostic reagents [16].
The purification and characterization of bacterial selenocysteine beta-lyase, an enzyme which specifically catalyzes the cleavage of L-selenocysteine to L-alanine, has been purified from Citrobacter freundii. The enzyme is monomeric with a molecular weight of ca. 64,000 and contains 1 mol of pyridoxal 5'-phosphate as a cofactor per mol of enzyme. The enzyme also catalyzes the alpha, beta elimination of beta-chloro-L-alanine to form NH3, pyruvate [15].
C. Freundii strains also carry a plasmid that encodes class 1 AmpC cephalosporinase. These enzymes can hydrolzye inactivate new cephamycins and cephalosporins [17].
Current Research
A small scale research concerning certain strains of C. freundii was done recently at the University of Tennessee, Knoxville. The importance of certain tetracycline and streptomycin resistance genes and class 1 integrons in C. freundii isolated from dairy farm soil and nondairy soils were evaluated. One strain of C. freundii extracted from dairy farm soils carried class 1 integrons with different inserted gene cassettes. Results of this small study suggested that the presence of multiple resistance genes and class 1 integrons in C. freundii in dairy farm soil may act as a reservoir of antimicrobial resistance genes and could play a role in the dissemination of these antimicrobial resistance genes to other commensal and indigenous microbial communities in soil. However, additional longer-term studies conducted in more locations are needed to support this hypothesis [12].
A second research concering C. freundii was done in order to devise a polymerase chain reaction (PCR) method that simultaneously uses three pairs of specific primers to detect genes of certain microbes (including C. freundii). The method included designing three primer pairs which were: SPVC-1 and SPVC-2, INVA-1 and INVA-2; and VIAB-1 and VIAB-2. PCR was performed using these three primers to identify 14 clinically important bacterial organisms. The following strains were quickly identified using the PCR: (1) C. freundii; (2) S. Typhi; and S. Paratyphi C; (3) S. Dublin (virulence antigen-positive); and (4) Salmonella serovars that harbor an spv-type virulence plasmid. Although this PCR method is new, with the advance of technology in the future this method can allow the identification of C. freundii in mammals immediately so that appropriate antibiotic treatment can be initiated without delay [13].
A third study concering C.freundii was done at the University of Barcelona, Spain. The mechanisms of resistance to fluoroquinolones in two Citrobacter freundii strains were studied. Both strains were isolated from the same patient. This study allowed partial characterisation of the acrA and acrB genes of this microorganism. Expression of genes in both strains was analysed using DNA microarrays for Escherichia coli. Nucleotide similarity between the partially sequenced acrA and acrB genes of C. freundii and E. coli was 80.7% and 85%, respectively. The acrA and acrB genes of C. freundii are similar to those in E. coli and their overexpression may play an important role in modulating the final minimum inhibitory concentration of fluoroquinolones [14].
A fourth study concerning C. freundii was done in Taiwan. A team of researchers isolated a diabetic patient that developed necrotizing fasciitis which was caused by C. freundii from an injury incited by a marine animal. Necrotizing fasciitis is an infection within the deeper layer of the skin and subcutaneous tissues. When treating the patient they took a sample from the fluid in the wound and found C. freundii. After three days of starting the antibiotic treatments with cefotaxmine and cefepmine, there was accumulation of subcutaneous abscesses. After 6, 10, 14, and 21 days of tending to the patient, some antibiotics were giving the patient some relief, but no long term recovery was reached. Cefotoaxime, cefepime, ciprofloxamin are among the antibiotics that were no match for C. freundii. The patient fully recovered after 42 days of ertapenem treatment. The researchers had isolated two colonies of C. freundii within 5 days of each other. As the isolates were grown they became immensely resistant to cefotaxime and cefepime. The reason why entrapenem worked against C. freundii is because is active against AmpC producing Enterobacteriaceae [21].
Antibiotic resistance
Citrobacter species are a common cause of nosocomial infections associated with patients that are undergoing prolonged hospital treatments. C. freundii has recently been reported to express resistance to broad-spectrum antibiotincs including piperacillin, piperacillintazobactam, vancomycin and cephalosporins. Isolation of ceftriaxone-resistant Citrobacter freundii (CRCF) has been associated with the overprescribed broad spectrum antibiotics. The emerging new CRCF strains could suggest induction or depression of resistance genes as well as elimination of competing organisms. CRCF has been mostly isolated from patients with significant comorbidities including AIDS, peripheral vascular disease, and cerebrovascular disease. The usage of fluoroquinolone has also been reported to have no affect against the isolation of CRCF [19].
Citrobacter freundii is also known to contain in its chromosome a gene coding for cephalosporinase. This enzyme hydrolyses −CO−NH− bond in the lactam ring of cephalosporins and cephamycis thus rendering the bacteria resistant to this type of antibiotics. However when exposed to new third generation cephems and cerbapenems, clinically isolated C.freundii showed sensitivity to those substances.
A small outbreak of C.freundii resistant to third generation cephems has been observed in the surgical ward of Nagoya University Hospital in patients that underwent surgical procedures. The C.freundii was isolated from patient's bile, wound gauze, feces, pus, and ascites. It was suggested that these new strains of C. freundii contained a plasmid encoding AmpC celphalosporinase but upon failure to transfer the cephems resistance from C. freundii to E.coli it was concluded that the enzyme must be encoded in the chromosome of C. freundii. Since C. freundi is associated with nosocomial infections caution to this new strains is recommended [20].
References
1. Wang JT,Chang SC, Chen YC, Luh KT. “Comparison of antimicrobial susceptibility of Citrobacter freundii isolates in two different time periods.” The Journal of Microbiology, Immunology and Infection. 2000 Dec; 33(4): 258-62.
2. Whalen JG, Mully TW, Enlgish JC 3rd. “Spontaneous Citrobacter freundii infection in an immunocompetent patient.” Archives of dermatology. 2007 Jan; 143(1): 124-5.
3. Puchenkova SG. “Enterobacteria in areas of water along the Crimean Coast.” Mikrobiolohichnyĭ zhurnal. 1996 Mar-Apr; 58(2): 3-7.
4.http://www.ncbi.nlm.nih.gov/sites/entrez?db=genomeprj&cmd=Retrieve&dopt=Overview&list_uids=13123
5. Ilya V. Manukhov, Daria V. Mamaeva, Sergei M. Rastorguev, Nicolai G. Faleev, Elena A. Morozova, Tatyana V. Demidkina, and Gennadii B. Zavilgelsky. “A Gene Encoding L-Methionine γ-Lyase Is Present in Enterobacteriaceae Family Genomes: Identification and Characterization of Citrobacter freundii L-Methionine γ-Lyase.” The Journal of Bacteriology. 2005 June; 187(11): 3889-3893.
6. Lindberg, Frederik; Westman, Lennart; Normark, Staffan. “Regulatory Components in Citrobacter freundii ampC β -lactamase Induction.” Proceedings of the National Academy of Sciences of the United States of America. 1985 July; 82(14): 4620-4624.
7. Tsukamoto K, Tachibana K, Yamazaki N, Ishii Y, Ujiie K, Nishida N, Sawai T. “Role of lysine-67 in the active site of class C beta-lactamase from Citrobacter freundii GN346.” European Journal of Biochemistry/FEBS. 1990 Feb 22; 188(1): 15-22.
8. Keevil CW, Hough JS, Cole JA. “Prototrophic growth of Citrobacter freundii and the biochemical basis for its apparent growth requirements in aerated media.” Journal of General Microbiology. 1997 Jan; 98(1): 273-6.
9. Marco Sánchez F, Turabian Fernández JL, Durán Pérez-Navarro A. “Fatal Citrobacter Freundii Bronchopneumonia Acquired in the Community in an Uncompromised Patient.” Revista Clínica Española. 1985 Apr; 176(6): 320
10. Julie L. Badger, Monique F. Stins, and Kwang Sik Kim. “Citrobacter freundii Invades and Replicates in Human Brain Microvascular Endothelial Cells.” Hinyokika kiyo. Acta Urologica Japonica. 1985 Jul; 31(7): 1159-70.
11. Drelichman V, Band JD. “Bacteremias due to Citrobacter diversus and Citrobacter freundii. Incidence, risk factors, and clinical outcome.” Archives of Internal Medicine. 1985 Oct; 145(10): 1808-10.
12. Srinivasan V, Nam HM, Sawant AA, Headrick SI, Nguyen LT, Oliver SP. “Distribution of Tetracycline and Streptomycin Resistance Genes and Class 1 Integrons in Enterobacteriaceae Isolated from Dairy and Nondairy Farm Soil.” Microbial Ecology. 2007 Aug 15.
13. Sánchez-Céspedes J, Vila J. “Partial characterisation of the acrAB locus in two Citrobacter freundii clinical isolates.” International Journal of Antimicrobial Agents. 2007 Sep; 30(3): 259-63.
14. Ciğerci IH, Korcan SE, Konuk M, Oztürk S. “Comparison of ALAD activities of Citrobacter and Pseudomonas strains and their usage as biomarker for Pb contamination.” Environmental Monitoring and Assessment. 2007 May 22.
15. Chocat P, Esaki N, Tanizawa K, Nakamura K, Tanaka H, Soda K. “Purification and characterization of selenocysteine beta-lyase from Citrobacter freundii.” Journal of Bacteriology. 1985 Aug; 163(2): 669-76.
16. Hillel S Levinson, Inga Mahler. “Phosphatase activity and lead resistance in Citrobacter freundii and Staphylococcus aureus.” FEMS Microbiology Letters. 1998 Apr; 161(1): 135-138.
17. Murakami K, Yoshida T. “Covalent binding of moxalactam to cephalosporinase of Citrobacter freundii.” Antimicrobial Agents and Chemotherapy. 1985 May; 27(5): 727-32.
18. Dervisoglu, E., Yegenaga, I., Yumuk, Z. “Citrobacter Freundii peritonitis and tunnel infection in a patient on continuous ambulatory peritoneal dialysis.” Journal of Medical Microbiology, 2008. Volume 57. p. 125-127.
19. Kim PW, Harris AD, Roghmann MC, Morris JG Jr, Strinivasan A, Perencevich EN. "Epidemiological risk factors for isolation of ceftriaxone-resistant versus -susceptible citrobacter freundii in hospitalized patients" Antimicrob Agents Chemother, 2003 Sep;47(9): p.2882-2887.
20. Nada T, Baba H, Kawamura K, Ohkura T, Torii K, Ohta M. "A small outbreak of third generation cephem-resistant Citrobacter freundii infection on a surgical ward" Jpn J Infect Dis. 2004 Aug;57(4): p.181-182.
21. Chuang, Y., Tseng, S.,Teng, L., Ho, Y., and Hsueh, P. "Emergence of cefotaxime resistance in Citrobactor freundii causing necrotizing fasciitis and osteomyelitis" Journal of Infection, 2006. Volume 53. p. e161-e163.
22.Badger, J., Stins, M., and Sik Kim, K. “Citrobacter freundii Invades and Replicates in Human Brain Microvascular Endothelial Cells” Infection and Immunity, 1999. Volume67. P. 4208-4215.
Edited by Sumaira Akbarzada, student of Rachel Larsen
Edited by Greg Vargas and Darren Zhen ,students of M Glogowskiat Loyola University
Edited by Gergana Grigorova and Michal Olszewski, students of M Glogowskiat Loyola University
Edited by Alex Kula and Joanna Aiken, students of M Glogowskiat Loyola University