Pneumocystis carinii: Difference between revisions
No edit summary |
|||
| (61 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | |||
{{Biorealm Genus}} | {{Biorealm Genus}} | ||
| Line 19: | Line 20: | ||
==Description and significance== | ==Description and significance== | ||
From the genus ''Pneumocystis'', ''Pneumocystis carinii'' | From the genus ''Pneumocystis'', ''Pneumocystis carinii'' is a fungal opportunistic pathogen which causes the disease Pneumocystis pneumonia (PCP) in immunocompromised mammalian hosts, specifically rats. ''Pneumocystis carinii'' was originally thought to be a developmental stage in the life cycle of ''Trypanosoma'' when it was first observed in 1909. The initial classification of ''Pneumocystis carinii'' as a protozoon was partly due to the life-cycle of the organism which involves a trophic and cyst form. In 1912 it was determined to be a different protozoon and named ''Pneumocystis carinii''. However, in 1988, the organism was reclassified as a member of the fungal kingdom after its small ribosomal RNA subunit was sequenced. The full genome has yet to be sequenced, but progress is currently underway, see genome structure (1). Although ''Pneumocystis carinii'' has yet to be grown in pure culture, aspects of its life-cycle have been observed within infected mammalian lung tissue. During its known life-cycle, ''P. carinii'' can be found as trophic and cystic forms (2). Still, much remains to be discovered about this organism. | ||
The HIV pandemic elevated interest in ''Pneumocystis carinii'' since it is the pathogen which caused Pneumocystis pneumonia and, consequently, a major cause of mortality in patients diagnosed with AIDS. Despite successes against ''P. carinii'' with trimethoprim with sulfamethoxazole there are signs that drug target sights may be mutating (1). | |||
It should be noted that both ''Pneumocystis carinii'' and ''Pneumocystis jirovecii'' currently refer to the same organism in medical literature with regards to PCP. ''P. jirovecii'' is the organism isolated from humans while ''P. carinii'' is found in rats (3). The two organisms cause PCP in their respective hosts. | |||
It should be noted that both ''Pneumocystis carinii'' and ''Pneumocystis jirovecii'' currently refer to the same organism in medical literature with regards to PCP. ''P. jirovecii'' is the organism isolated from humans while ''P. carinii'' is found in rats(3). The two organisms cause PCP in their respective hosts. | |||
==Genome structure== | ==Genome structure== | ||
Sequencing of the Pneumocystis carinii is currently in progress and knowledge about the gnome has been advanced by the [http://pgp.cchmc.org/ ''Pneumocystis'' Genome Project] and the [http://www.sanger.ac.uk/Projects/P_carinii/ Wellcome Trust Sanger Institute ''Pneumocystis carnii'' telomeric clone sequencing project]. | Sequencing of the ''Pneumocystis carinii'' genome is currently in progress and knowledge about the gnome has been advanced by groups such as the [http://pgp.cchmc.org/ ''Pneumocystis'' Genome Project] and the [http://www.sanger.ac.uk/Projects/P_carinii/ Wellcome Trust Sanger Institute ''Pneumocystis carnii'' telomeric clone sequencing project] in conjunction with the [http://www.ncbi.nlm.nih.gov/sites/entrez?db=genomeprj&cmd=Retrieve&dopt=Overview&list_uids=125 University of Cincinnati, the University of Georgia, and the University of Kentucky]. Information from these programs has not been made directly available to the public at this point in time. | ||
The ''P. carinii'' genome | The ''P. carinii'' genome consists of 15 chromosomes which are from 300 kb to 700 kb in size. On these 15 chromosomes there are approximately 8 million base pairs of DNA. One feature of the ''P. carinii'' genome is that the organism has a high concentration of adenine and thymine bases (65% of the total base pairs). The genome has introns which contain 38 to 424 base pairs and there have been up to nine introns observed per gene. The yeast ''Schizosaccharomyces pombe'' has been used as a homologue for the expressed sequenced tags (ESTs) found within the ''P. carinii genome'' (4). Completion of genomic sequence will prove invaluable in unlocking the biology of ''Pneumocystis carinii'' beyond what has been determined through the study of infected mammalian lung tissue. | ||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
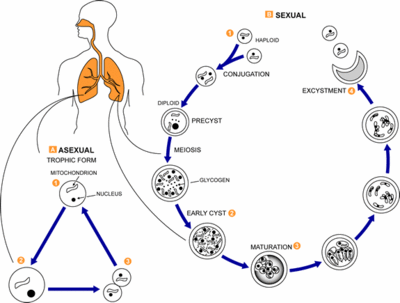
[[Image:Pneumocystis_LifeCycle.gif|thumb|Pneumocystis Life Cycle|400px|right|Life cycle of ''Pneumocystis carinii''. From [http://www.dpd.cdc.gov/dpdx/HTML/Pneumocystis.htm Centers for Disease Control and Prevention Division of Parasitic Diseases (DPDx).]]] | |||
Like other fungi, ''Pneumocystis carinii'' has chitin in its cell wall. The cyst form also contains β-1,3-glucan within its cell wall. This structural protein may help elicit an inflammatory response from the host and, subsequently, may be one of the factors contributing to damage of the alveolar epithelium. ''P. carinii'' uses mannose carbohydrates to adhere to the host (1). These surface antigens are unique to ''P. carinii'' and might provide a means of evading the host immune system. | |||
''Pneumocystis carinii'' exists within the host in two forms: the trophic and the cyst. The trophozoites average a diameter of 1-4µm while the cysts are 8-10µm. The trophic forms of ''P. carinii'' may reproduce asexually via binary fission while cysts are able to reproduce sexually after trophozoites conjugate (2). The cysts develop in three stages with increasing amounts of intracystic nuclei in each stage. The early cyst stage contains two intracystic nuclei while the intermediate stage has four and the late precysts stage holds eight. The mature cysts then release the trophic form of ''P. carinii'' as they burst. These observations on the life cycle have been made without the organism growing in a pure culture and contradict the life cycles of other ascomyceteous fungi (1). The life cycle is similar to those observed in protozoa and led to the initial classification of ''P. carinii'' as a protozoon. | |||
The metabolism of lipids by ''Pneumocystis carinii'' has been unlocked by the use of radiolabeled precursors in purified samples of the organism. ''P. carinii'' is capable of incorporating ethanolamine and choline precursors into phospholipid head groups. Additionally, it was found that the organism is capable of metabolizing PE to PC by the methylation of the glycerophospholipid head group. Experiments using radiolabeled serine and inositol have established that ''P. carinii'' might be able to convert PS to PI (5). | |||
==Ecology== | ==Ecology== | ||
''P. carinii'' | Thus far, ''P. carinii'' is found in the lung tissue of immunocompetent and immunocompromised hosts. The organism adheres specifically to the epithelium of the alveoli within the lung. Only hosts with compromised immune systems are at risk of developing PCP from ''P. carinii'' while a normal host is able to mount an adequate immune response to contain ''P. carinii''. | ||
Fragments of DNA identical to DNA sequences found in ''P. carinii'' have been found in ambient air( | Fragments of DNA identical to DNA sequences found in ''P. carinii'' have been found in ambient air(6) and pond water (7). Even though ''P. carinii'' nucleic acids have been found outside of the host environment, there is no definitive evidence to support an ecological niche for ''Pneumocystis carinii'' outside of a host environment. | ||
==Pathology== | ==Pathology== | ||
When ''Pneumocystis carinii'' enters the lungs of a host with a normal immune system the pathogenic fungi does not cause Pneumocystis pneumonia. ''P. carinii'' fails to cause PCP in a host with a normal immune system because it is an opportunistic pathogen which is kept in check by a host with a fully functional immune response. The onset of PCP is the result an infection, within a host that has a compromised immune system, because the organism is able to proliferate. Uninhibited growth results in pathogenesis. Symptoms of PCP include: shortness of breath, non-productive cough, fever, malaise, and chest pain. An infection is fatal, if left untreated, because the trophic form of ''P. carinii'' binds to the type I pneumocytes and prevents gas to be exchanged between capillaries and the alveoli. Untreated inhibition of gas exchange in the host leads to poor oxygenation, hypoxemia and death. Treatments include the use of trimethoprim with sulfamethoxazole or pentamidine (2). | |||
== | ==Current Research== | ||
'''"Transcriptome of Pneumocystis carinii during Fulminate Infection: Carbohydrate Metabolism and the Concept of a Compatible Parasite"''' (8) | |||
ESTs were taken from the poly(A) tails of ''P. carinii'' messenger RNA to determine what types of genes were expressed during an infection. This strategy does not provide a complete picture of all the genes that may be expressed by ''P. carinii'' since not all genes are expressed at one time. The greater majority of the transcripts observed were part of metabolic processes and stress responses. Nutrient limitations on the organisms increase after they have significantly compromised the oxygen flow in the alveoli. The CON7 gene, which is associated with sporogenesis in fungi, was found to be expressed in abundance. This suggests that nutrient deprivation can cause ''P. carinii'' to enter the sexual reproductive stage of its life cycle. Thus, thus the oxygen supplied by the host's biology is possibly the trigger for the organism to move on to the next stage in its life cycle and the reason why it has not been seen outside of a host. Understanding what genes are being expressed during an infection is an important first step and determining drug targets unique to ''P. carinii'' in order to minimize collateral damage to the host. | |||
'''"Parameters Determining the Relative Efficacy of Hydroxy-naphthoquinone Inhibitors of the Cytochrome bc1 Complex"''' (9) | |||
With the rise in drug resistances in ''P. carinii'' it is very important to find ways to regain antimicrobial supremacy with the development of new drugs. Atovaquone®, a hydroxy-naphthoquinone is currently being used to competitively inhibit the cytochrome ''bc1'' complex that binds the ubiquinol oxidation site between cytochrome ''b'' and the iron-sulfur protein. Yeast and bovine versions of the complex were used to test modified hydroxy-naphthoquinones against the mutated cytochrome ''b'' gene in ''P. carinii''. A steroisomer was developed and proved successful against the mutant. However, it could be quickly broken down by the human body. Despite the shortcomings experienced the new steroisomer represents a new lead to be used in the fight against resistance in ''P. carinii''. | |||
'''"Exploiting Amoeboid and Non-Vertebrate Animal Model Systems to Study the Virulence of Human Pathogenic Fungi"''' (10) | |||
Traditionally mammalian models have been used to study how the immune system combats fungal pathogens like ''P. carinii''. Non-vertebrate models can be used to the same effect to study the innate immune system. Innate immune responses are conserved across phyla. By using non-vertebrates in addition to the traditional models, researchers are able to study the effects of fungal pathogens in a wider range of host environments. Several advantages found in using non-vertebrate models are that non-vertebrate models are less complex in their biology than mammalian models and are a relatively inexpensive, in comparison to mammalian models, option. These advantages could help researchers by providing alternative host models for fungal pathogens, at a more affordable cost, to augment existing models. The greater accessibility and variety of host choices would be better suited to compete with the evolution ''P. carinii''. | |||
==References== | ==References== | ||
Edited by Daniel Spisak of [mailto:ralarsen@ucsd.edu Rachel Larsen] | 1. [http://www.nature.com/nrmicro/journal/v5/n4/full/nrmicro1621.html Thomas, Charles F. Jr, Limper, Andrew H. "Current insights into the biology and pathogenesis of ''Pneumocystis'' pneumonia". ''Nature Reviews Microbiology''. April 2007. Volume 5. p. 298-308.] | ||
2. [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6TD0-4C1FD61-1&_user=4429&_coverDate=05%2F31%2F2004&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000059602&_version=1&_urlVersion=0&_userid=4429&md5=8371790c3a2534bb87df74e47a011c7e Cushion, Melanie T. "''Pneumocystis'': unraveling the cloak of obscurity". ''Trends in Microbiology''. May 2004. Volume 12. p. 243-249.] | |||
3. [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6W8X-4MKW6BR-2&_user=4429&_coverDate=01%2F31%2F2007&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000059602&_version=1&_urlVersion=0&_userid=4429&md5=e4c932b71c375fd86452286e2b0bfa4d Hawksworth, David L. "Responsibility in naming pathogens: the case of ''Pneumocystis jirovecii'', the causal agent of pneumocystis pneumonia". ''The Lancet Infectious Diseases''. February 2007. Volume 7. p. 92.] | |||
4. [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=15965256 Keely S.P., Renauld H., Wakefield A.E., Cushion M.T., Smulian A.G., Fosker N., Fraser A., Harris D., Murphy L., Price C., et al. "Gene arrays at ''Pneumocystis carinii'' telomeres". ''Genetics''. August 2005. Volume 105. p. 1589–1600.] | |||
5. [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=9457427#id2622059 Kaneshiro, Edna S. "The Lipids of ''Pneumocystis carinii''". ''Clinical Microbiology Reviews''. January 1998. Volume 11. p. 27-41.] | |||
6. [http://jcm.asm.org/cgi/content/abstract/34/7/1754 Wakefield , A.E. "DNA sequences identical to Pneumocystis carinii f. sp. carinii and Pneumocystis carinii f. sp. hominis in samples of air spora". ''Journal of Clinical Microbiology''. July 1996. Volume 34. p. 1754-1759.] | |||
7. [http://www.blackwell-synergy.com/doi/pdf/10.1111/j.1550-7408.1997.tb05752.x Casanova-Cardiel, Luis, Leibowitz, Michael J. "Presence of ''Pneumocystis carinii'' DNA in Pond Water". ''The Journal of Eukaryotic Microbiology''. 1997. Volume 44. p. 28s.] | |||
8. [http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1855432#id2650713 Cushion, Melanie T., Smulian, A. George., et al. "Transcriptome of ''Pneumocystis carinii'' during Fulminate Infection: Carbohydrate Metabolism and the Concept of a Compatible Parasite". ''PLoS ONE''. May 2007. Volume 2. p. e423.] | |||
9. [http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1939967 Kessl, Jacques J., Moskalev, Nikolai V., Gribble, Gordon W., et al. "Parameters Determining the Relative Efficacy of Hydroxy-naphthoquinone Inhibitors of the Cytochrome ''bc1'' Complex". ''Biochim Biophys Acta''. February 2007. Volume 1767. p. 319-326.] | |||
10. [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1933451 Finlay, B. Brett, ed. "Exploiting Amoeboid and Non-Vertebrate Animal Model Systems to Study the Virulence of Human Pathogenic Fungi". ''PLoS Pathog''. July 2007. Volume 3. p. e101.] | |||
Edited by Daniel Spisak, student of [mailto:ralarsen@ucsd.edu Rachel Larsen] | |||
Edited by KLB | |||
Latest revision as of 03:29, 20 August 2010
A Microbial Biorealm page on the genus Pneumocystis carinii
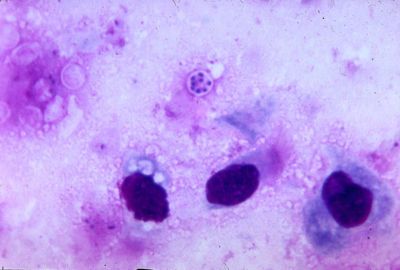
Classification
Higher order taxa
Eukaryota; Fungi; Dikarya; Ascomycota; Taphrinomycotina; Pneumocystidomycetes; Pneumocystidaceae; Pneumocystis
Species
Pneumocystis carinii
|
NCBI: Taxonomy |
Description and significance
From the genus Pneumocystis, Pneumocystis carinii is a fungal opportunistic pathogen which causes the disease Pneumocystis pneumonia (PCP) in immunocompromised mammalian hosts, specifically rats. Pneumocystis carinii was originally thought to be a developmental stage in the life cycle of Trypanosoma when it was first observed in 1909. The initial classification of Pneumocystis carinii as a protozoon was partly due to the life-cycle of the organism which involves a trophic and cyst form. In 1912 it was determined to be a different protozoon and named Pneumocystis carinii. However, in 1988, the organism was reclassified as a member of the fungal kingdom after its small ribosomal RNA subunit was sequenced. The full genome has yet to be sequenced, but progress is currently underway, see genome structure (1). Although Pneumocystis carinii has yet to be grown in pure culture, aspects of its life-cycle have been observed within infected mammalian lung tissue. During its known life-cycle, P. carinii can be found as trophic and cystic forms (2). Still, much remains to be discovered about this organism.
The HIV pandemic elevated interest in Pneumocystis carinii since it is the pathogen which caused Pneumocystis pneumonia and, consequently, a major cause of mortality in patients diagnosed with AIDS. Despite successes against P. carinii with trimethoprim with sulfamethoxazole there are signs that drug target sights may be mutating (1).
It should be noted that both Pneumocystis carinii and Pneumocystis jirovecii currently refer to the same organism in medical literature with regards to PCP. P. jirovecii is the organism isolated from humans while P. carinii is found in rats (3). The two organisms cause PCP in their respective hosts.
Genome structure
Sequencing of the Pneumocystis carinii genome is currently in progress and knowledge about the gnome has been advanced by groups such as the Pneumocystis Genome Project and the Wellcome Trust Sanger Institute Pneumocystis carnii telomeric clone sequencing project in conjunction with the University of Cincinnati, the University of Georgia, and the University of Kentucky. Information from these programs has not been made directly available to the public at this point in time.
The P. carinii genome consists of 15 chromosomes which are from 300 kb to 700 kb in size. On these 15 chromosomes there are approximately 8 million base pairs of DNA. One feature of the P. carinii genome is that the organism has a high concentration of adenine and thymine bases (65% of the total base pairs). The genome has introns which contain 38 to 424 base pairs and there have been up to nine introns observed per gene. The yeast Schizosaccharomyces pombe has been used as a homologue for the expressed sequenced tags (ESTs) found within the P. carinii genome (4). Completion of genomic sequence will prove invaluable in unlocking the biology of Pneumocystis carinii beyond what has been determined through the study of infected mammalian lung tissue.
Cell structure and metabolism
Like other fungi, Pneumocystis carinii has chitin in its cell wall. The cyst form also contains β-1,3-glucan within its cell wall. This structural protein may help elicit an inflammatory response from the host and, subsequently, may be one of the factors contributing to damage of the alveolar epithelium. P. carinii uses mannose carbohydrates to adhere to the host (1). These surface antigens are unique to P. carinii and might provide a means of evading the host immune system.
Pneumocystis carinii exists within the host in two forms: the trophic and the cyst. The trophozoites average a diameter of 1-4µm while the cysts are 8-10µm. The trophic forms of P. carinii may reproduce asexually via binary fission while cysts are able to reproduce sexually after trophozoites conjugate (2). The cysts develop in three stages with increasing amounts of intracystic nuclei in each stage. The early cyst stage contains two intracystic nuclei while the intermediate stage has four and the late precysts stage holds eight. The mature cysts then release the trophic form of P. carinii as they burst. These observations on the life cycle have been made without the organism growing in a pure culture and contradict the life cycles of other ascomyceteous fungi (1). The life cycle is similar to those observed in protozoa and led to the initial classification of P. carinii as a protozoon.
The metabolism of lipids by Pneumocystis carinii has been unlocked by the use of radiolabeled precursors in purified samples of the organism. P. carinii is capable of incorporating ethanolamine and choline precursors into phospholipid head groups. Additionally, it was found that the organism is capable of metabolizing PE to PC by the methylation of the glycerophospholipid head group. Experiments using radiolabeled serine and inositol have established that P. carinii might be able to convert PS to PI (5).
Ecology
Thus far, P. carinii is found in the lung tissue of immunocompetent and immunocompromised hosts. The organism adheres specifically to the epithelium of the alveoli within the lung. Only hosts with compromised immune systems are at risk of developing PCP from P. carinii while a normal host is able to mount an adequate immune response to contain P. carinii.
Fragments of DNA identical to DNA sequences found in P. carinii have been found in ambient air(6) and pond water (7). Even though P. carinii nucleic acids have been found outside of the host environment, there is no definitive evidence to support an ecological niche for Pneumocystis carinii outside of a host environment.
Pathology
When Pneumocystis carinii enters the lungs of a host with a normal immune system the pathogenic fungi does not cause Pneumocystis pneumonia. P. carinii fails to cause PCP in a host with a normal immune system because it is an opportunistic pathogen which is kept in check by a host with a fully functional immune response. The onset of PCP is the result an infection, within a host that has a compromised immune system, because the organism is able to proliferate. Uninhibited growth results in pathogenesis. Symptoms of PCP include: shortness of breath, non-productive cough, fever, malaise, and chest pain. An infection is fatal, if left untreated, because the trophic form of P. carinii binds to the type I pneumocytes and prevents gas to be exchanged between capillaries and the alveoli. Untreated inhibition of gas exchange in the host leads to poor oxygenation, hypoxemia and death. Treatments include the use of trimethoprim with sulfamethoxazole or pentamidine (2).
Current Research
"Transcriptome of Pneumocystis carinii during Fulminate Infection: Carbohydrate Metabolism and the Concept of a Compatible Parasite" (8)
ESTs were taken from the poly(A) tails of P. carinii messenger RNA to determine what types of genes were expressed during an infection. This strategy does not provide a complete picture of all the genes that may be expressed by P. carinii since not all genes are expressed at one time. The greater majority of the transcripts observed were part of metabolic processes and stress responses. Nutrient limitations on the organisms increase after they have significantly compromised the oxygen flow in the alveoli. The CON7 gene, which is associated with sporogenesis in fungi, was found to be expressed in abundance. This suggests that nutrient deprivation can cause P. carinii to enter the sexual reproductive stage of its life cycle. Thus, thus the oxygen supplied by the host's biology is possibly the trigger for the organism to move on to the next stage in its life cycle and the reason why it has not been seen outside of a host. Understanding what genes are being expressed during an infection is an important first step and determining drug targets unique to P. carinii in order to minimize collateral damage to the host.
"Parameters Determining the Relative Efficacy of Hydroxy-naphthoquinone Inhibitors of the Cytochrome bc1 Complex" (9)
With the rise in drug resistances in P. carinii it is very important to find ways to regain antimicrobial supremacy with the development of new drugs. Atovaquone®, a hydroxy-naphthoquinone is currently being used to competitively inhibit the cytochrome bc1 complex that binds the ubiquinol oxidation site between cytochrome b and the iron-sulfur protein. Yeast and bovine versions of the complex were used to test modified hydroxy-naphthoquinones against the mutated cytochrome b gene in P. carinii. A steroisomer was developed and proved successful against the mutant. However, it could be quickly broken down by the human body. Despite the shortcomings experienced the new steroisomer represents a new lead to be used in the fight against resistance in P. carinii.
"Exploiting Amoeboid and Non-Vertebrate Animal Model Systems to Study the Virulence of Human Pathogenic Fungi" (10)
Traditionally mammalian models have been used to study how the immune system combats fungal pathogens like P. carinii. Non-vertebrate models can be used to the same effect to study the innate immune system. Innate immune responses are conserved across phyla. By using non-vertebrates in addition to the traditional models, researchers are able to study the effects of fungal pathogens in a wider range of host environments. Several advantages found in using non-vertebrate models are that non-vertebrate models are less complex in their biology than mammalian models and are a relatively inexpensive, in comparison to mammalian models, option. These advantages could help researchers by providing alternative host models for fungal pathogens, at a more affordable cost, to augment existing models. The greater accessibility and variety of host choices would be better suited to compete with the evolution P. carinii.
References
Edited by Daniel Spisak, student of Rachel Larsen
Edited by KLB
