Epulopiscium fishelsoni: Difference between revisions
No edit summary |
Slonczewski (talk | contribs) No edit summary |
||
| (30 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | |||
{{Biorealm Genus}} | {{Biorealm Genus}} | ||
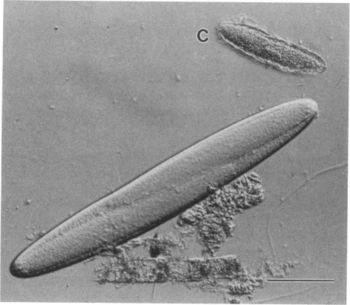
[[Image: | [[Image:E.fish.JPG|thumb|350px|right|Light micrograph of a large epulo and a ciliated eukaryote (C). [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC208246/pdf/jbacter00107-0127.pdf Clements and Bullivant, Copyright © 1991, American Society for Microbiology]]] | ||
Copyright © | |||
==Classification== | ==Classification== | ||
| Line 30: | Line 28: | ||
==Description and significance== | ==Description and significance== | ||
Epulopiscium was first discovered in 1985 by Lev Fishelson. He found them inside the intestines of a brown surgeonfish and was initially classified as a protist because of its large size. ''Epulopiscium fishelsoni'' was later discovered by Linn Montgomery and in 1993 Norman Pace performed an rRNA analysis which confirmed that it was actually a bacteria. At the time ''E. fishelsoni'' was the largest known bacteria, ranging from a length of ~30 to >600μm and a volume of >2,000-fold. The cell size also varies more than in any other bacteria [2]. Cellular or molecular mechanisms that may support and control such exceptional variability in the sizes of ''E. fishelsoni'' are unclear. However, the cells are highly mobile, vary in mean size and structure during a 24 hour period, and affect the pH of the host’s gut fluids differentially during day and night. ''E. fishelsoni'' is a gram-positive bacterium that has an explicit symbiotic relationship with its surgeonfish hosts [2]. | |||
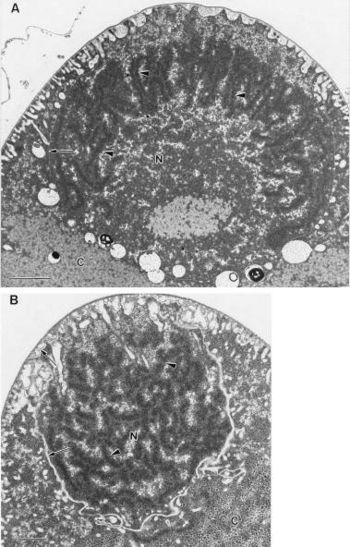
[[Image:Tileshop.fcgi.jpg|thumb|350px|right|Transmission electron micrographs of compact nucleoids of ''E. fishelsoni''. Copyright © 1998, American Society for Microbiology]] | |||
==Genome structure== | ==Genome structure== | ||
The size of a unit genome in ''E. fishelsoni'' is not known, but copy number in large cells appears to be very great. There are large quantities of DNA in large cells, there is localization of DNA-specific stains to nucleoids, and fluorescence ratios indicate condensation of DNA when electron micrographs show darkly staining, elongate structures in the nucleoids [2]. | |||
Many copies of a unit genome may support the growth, mobility, and active metabolism for these giant bacteria. Multiple copies of an entire genome would also support rapid production of daughter cells by uncoupling potentially rate-limiting DNA replication from DNA-subdivision and other cell division events [2]. | |||
Unlike most bacteria, ''E. fishelsoni'' reproduces exclusively through an unusual form of sporulation. In this process multiple daughter cells are grown inside of the parent cell until the daughter cells eventually become fully developed and then they burst through the cell wall. Although sporulation is a common process among other bacteria, it is a desperation measure that is performed by overcrowding or starvation, instead of a standard process of reproduction. The daughter cells in standard sporulation are usually dormant, however, the new daughter cells of ''E. fishelsoni'' are active and mobile [3]. | |||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
''E. fishelsoni'' has a layer of flagella around its surface. It possesses bacterial type flagella rather than eukaryote type cilia. When viewed in a light microscope video the flagella layer is vibrating in a wave-like motion which is what makes the cell swim. Beneath the outer surface is a peripheral layer of rolled up membranes, followed by a dense region with large, lightly stained inclusions. In the central region of the cell there are often membrane-bound daughter cells that is involved in the reproductive process. When the daughter cells completely develop, they emerge through an oblong split in the envelope of the mother cell [1]. | |||
==Ecology== | ==Ecology== | ||
General aspects of the ''E. fishelsoni'' life cycle correspond with activities of the host. During the day, the cell increases in size and nucleoids migrate to the poles of the cell and begin to elongate when the host is most active. The hosts are eating and their gut is completely filled with algae and food materials. As the day goes on, the average length of the cells increase, until the nucleoids make up a large percentage of the parent cell volume, and the sporulation process begins in the late afternoons and evenings. During the day as well, pH of gut fluids is suppressed in areas where ''E. fishelsoni'' occur in abundance. The total oppostite occurs at night. When the hosts are inactive and in their shelter reefs, the size of the cells are smaller, and there is no supression of pH in the gut [2]. | |||
==Pathology== | ==Pathology== | ||
''E. fishelsoni'' is not known to cause any disease or be harmful to anything. It has a symbiotic relationship with surgeonfish which seems to be mutually beneficial. In fact, ''E. fishelsoni'' can not be cultured outside of its host and can only be found in the gut of that family of fish. | |||
==Application to Biotechnology== | ==Application to Biotechnology== | ||
''E. fishelsoni'' does not produce any useful compound or enzymes that can be used for biotechnology. All that is known is that this bacteria helps regulate the pH in the gut of its host. This regulation is cycled throughout the day and is controlled based on the surgeonfish's activity. | |||
==Current Research== | ==Current Research== | ||
There are no ongoing | There are currently no ongoing nor recent research being done about ''E. fishelsoni''. After its discovery in 1985, thorough examinations of its structure and research about its reproduction process in the 90's, there is not much left that we don't know about this bacteria. There are no signs or indications of future research projects for ''E. fishelsoni''. | ||
==References== | ==References== | ||
| Line 63: | Line 71: | ||
Edited by student of [mailto:ralarsen@ucsd.edu Rachel Larsen] | Edited by student of [mailto:ralarsen@ucsd.edu Rachel Larsen] | ||
Edited by KLB | |||
Latest revision as of 16:18, 23 February 2012
A Microbial Biorealm page on the genus Epulopiscium fishelsoni
Classification
Higher order taxa
Kingdom: Bacteria
Phylum: Firmicutes
Class: Clostridia
Order: Clostridiales
Genus: Epulopiscium
Species
|
NCBI: Taxonomy |
Epulopiscium fishelsoni
Description and significance
Epulopiscium was first discovered in 1985 by Lev Fishelson. He found them inside the intestines of a brown surgeonfish and was initially classified as a protist because of its large size. Epulopiscium fishelsoni was later discovered by Linn Montgomery and in 1993 Norman Pace performed an rRNA analysis which confirmed that it was actually a bacteria. At the time E. fishelsoni was the largest known bacteria, ranging from a length of ~30 to >600μm and a volume of >2,000-fold. The cell size also varies more than in any other bacteria [2]. Cellular or molecular mechanisms that may support and control such exceptional variability in the sizes of E. fishelsoni are unclear. However, the cells are highly mobile, vary in mean size and structure during a 24 hour period, and affect the pH of the host’s gut fluids differentially during day and night. E. fishelsoni is a gram-positive bacterium that has an explicit symbiotic relationship with its surgeonfish hosts [2].
Genome structure
The size of a unit genome in E. fishelsoni is not known, but copy number in large cells appears to be very great. There are large quantities of DNA in large cells, there is localization of DNA-specific stains to nucleoids, and fluorescence ratios indicate condensation of DNA when electron micrographs show darkly staining, elongate structures in the nucleoids [2].
Many copies of a unit genome may support the growth, mobility, and active metabolism for these giant bacteria. Multiple copies of an entire genome would also support rapid production of daughter cells by uncoupling potentially rate-limiting DNA replication from DNA-subdivision and other cell division events [2].
Unlike most bacteria, E. fishelsoni reproduces exclusively through an unusual form of sporulation. In this process multiple daughter cells are grown inside of the parent cell until the daughter cells eventually become fully developed and then they burst through the cell wall. Although sporulation is a common process among other bacteria, it is a desperation measure that is performed by overcrowding or starvation, instead of a standard process of reproduction. The daughter cells in standard sporulation are usually dormant, however, the new daughter cells of E. fishelsoni are active and mobile [3].
Cell structure and metabolism
E. fishelsoni has a layer of flagella around its surface. It possesses bacterial type flagella rather than eukaryote type cilia. When viewed in a light microscope video the flagella layer is vibrating in a wave-like motion which is what makes the cell swim. Beneath the outer surface is a peripheral layer of rolled up membranes, followed by a dense region with large, lightly stained inclusions. In the central region of the cell there are often membrane-bound daughter cells that is involved in the reproductive process. When the daughter cells completely develop, they emerge through an oblong split in the envelope of the mother cell [1].
Ecology
General aspects of the E. fishelsoni life cycle correspond with activities of the host. During the day, the cell increases in size and nucleoids migrate to the poles of the cell and begin to elongate when the host is most active. The hosts are eating and their gut is completely filled with algae and food materials. As the day goes on, the average length of the cells increase, until the nucleoids make up a large percentage of the parent cell volume, and the sporulation process begins in the late afternoons and evenings. During the day as well, pH of gut fluids is suppressed in areas where E. fishelsoni occur in abundance. The total oppostite occurs at night. When the hosts are inactive and in their shelter reefs, the size of the cells are smaller, and there is no supression of pH in the gut [2].
Pathology
E. fishelsoni is not known to cause any disease or be harmful to anything. It has a symbiotic relationship with surgeonfish which seems to be mutually beneficial. In fact, E. fishelsoni can not be cultured outside of its host and can only be found in the gut of that family of fish.
Application to Biotechnology
E. fishelsoni does not produce any useful compound or enzymes that can be used for biotechnology. All that is known is that this bacteria helps regulate the pH in the gut of its host. This regulation is cycled throughout the day and is controlled based on the surgeonfish's activity.
Current Research
There are currently no ongoing nor recent research being done about E. fishelsoni. After its discovery in 1985, thorough examinations of its structure and research about its reproduction process in the 90's, there is not much left that we don't know about this bacteria. There are no signs or indications of future research projects for E. fishelsoni.
References
Edited by student of Rachel Larsen
Edited by KLB