Thermos Thermophilus: Difference between revisions
No edit summary |
|||
| (3 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | |||
{{Biorealm Genus}} | {{Biorealm Genus}} | ||
| Line 43: | Line 44: | ||
==Ecology== | ==Ecology== | ||
{| border="1" | {| border="1" | ||
!Most Thermus Thermophilus can be found in various geothermal environment through out the earth such as hot spring, undersea volcanic thermal vents. Thermophiles can live in acidic condition as low as pH 3.4 to very basic alkaline environment such as pH 9. The ability that allows them to survive these environments is all encoded in their gene and protein structure.!! Thermus Environment [[Image:Grand_prismatic_spring.jpg| | !Most Thermus Thermophilus can be found in various geothermal environment through out the earth such as hot spring, undersea volcanic thermal vents. Thermophiles can live in acidic condition as low as pH 3.4 to very basic alkaline environment such as pH 9. The ability that allows them to survive these environments is all encoded in their gene and protein structure.!! Thermus Environment [[Image:Grand_prismatic_spring.jpg|thumb|A picture taken from hot spring where thermus species spawn|400px|right]] | ||
|- | |- | ||
|} | |} | ||
Latest revision as of 03:37, 20 August 2010
A Microbial Biorealm page on the genus Thermos Thermophilus
Classification
Higher order taxa
Higher order taxa: Bacteria (Domain): Deinococcus-Thermus (Phylum): Deinococci (Class): Thermales (Order): Thermacaea (Family): Thermus (Genus) [1] link to find]
Species
Thermophilus (Species): Strain (HB27, HB8)
|
NCBI: Taxonomy |
Genus species
Description and significance
| Thermus Thermophilus is a Gram-negative bacterium that was isolated in 1971, Japan. They spawn in thermal spring ranging from 50-82C. The biological machines from these organisms have a higher stability than other organisms due to the environment that they have to live in. In general, thermophiles are anaerobes that can live in hot environment with low oxygen solubility due to the temperature with the exception of thermus, they are aerobic chemorganotroph. Thermus Thermophilus contains two strains, HB8 and HB27; both were found in Japan’s thermal environment with optimum environment 68C and the pH 7.0. The HB8 strain can live in either anaerobe and aerobe; where as the HB27 can only strive in aerobe environment. HB8 survive anaerobeically in the presence of nitrate through nitrate reductase production. However the HB27 was unable to growth in the same environment as the HB8 due to the inability to produce nitrate reductase. | Thermus Thermophilus |
|---|
Genome structure
The Thermus Thermophilus bacterium contain Circular DNA and a megaplasmid. The DNA has 1,894,877 base pair where 1476627 base pair (69.40%) are G+C content. The high percentage of G+C content allow the bacterium to strive in extreme thermo environment where it's own genetic information would not be denatured by the surrounding environment. Also, the megaplasmid contain 232,605 nucleotides with 69% G+C content. In addition, it contained a total of 2210 protein encoding genes and 53 RNA genes. (2)
|
NCBI: Genome structure |
Cell structure and metabolism
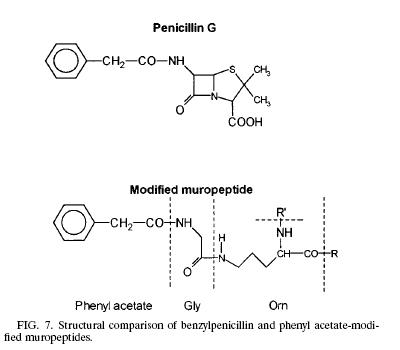
| Like any other Gram negative bacteria, Thermus Thermophilus composed of an outer membrane, mainly phospholipids and lipopolysaccharides, which made it ineffective to hold the crystal violet color during gram stain; a thin layer of peptidoglycan covering the plasma membrane and a cytoplasmic membrane. The peptidoglycan (murein) is responsible for the cell’s rigid structure. There are a total of 29 muropeptides composed of more than 85% of the total murein in the organism. Scientists dissected the composition of the Thermus Thermophilus murein and found the presence of Ala, Glu, Gly, Orn, N-acetyl glucosamine, and N-acetylmuramic. In addition to the amino acid and sugar mentioned above, T. Thermophilus also contains phenyl acetic acid at the N terminal of Glysine. The presence of phenyl acetic acid in the muropeptides is 23.7% relative to the total muropeptides. The process of how and why the phenyl acetic acid incorporated into the muropeptides is still unknown but scientists think that the aromatic phenyl ring could be facilitating the interaction between the noncovalent and hydrophobic molecules from the surrounding environment. Other hypothesis also arises linking the phenyl acetic acid to the structure of penicillin.(1) | Modified Muropeptide & Penicillin |
|---|
Ecology
| Most Thermus Thermophilus can be found in various geothermal environment through out the earth such as hot spring, undersea volcanic thermal vents. Thermophiles can live in acidic condition as low as pH 3.4 to very basic alkaline environment such as pH 9. The ability that allows them to survive these environments is all encoded in their gene and protein structure. | Thermus Environment |
|---|
Application to Biotechnology
The Thermus Thermosphilus’ enzyme is very stable and is the major topic for biotechnology. All of the enzymes display a much higher stability and resistance to denaturation from heat and chemical reagent than the mesophilic homologous, which make it a very appealing for industrial process. One of the enzymes from the thermus species had already been applied in scientific research and industrial application is the rTtH DNA polymerase, use in PCR. The rTtH DNA polymerase has an optimum temperature of 70-80C and in the presence of Magnesium 2+, it does show the effect of reverse transcriptase.(7)
Recent studied showed that T. Thermophilus DNA mismatch repair mechanism (MutS gene) can be transfer to other bacteria (E.coli) and still function normally. The MutS gene then undergoes mass production in the E.coli bacteria and purified. Nearly half of the MutS gene was stable in pH of 1.5 to as basic as 12 at room temperature and still stable upto as high as 80C. (5)
Current Research
Many researches on thermophilic organisms are currently taking place today. One of the researches was based on the production of polyester from the Thermus Thermophilus bacterium to make Polyhydroxylalkanoate (PHA) by the University of Greece. The substrate that was used as the carbon source to grow the bacterium was sodium gluconate or sodium octanoate. The result yielded PHA synthesis from 30-40% of the cellular dry weight(3). Other researchers tried to synthesize vitamin B12 from the Uroporphyrinogen-III C-methyltransferase, a multifunctional protein that responsible for two of the eight S-adenosyl-methionine-dependent methylations of the corrin ring during vitamin B(12) synthesis.(4)
References
1. JOSE CARLOS QUINTELA, ERNST P.,GUNTER A.,VICENTE A.,MIGUEL A. DE P. "Structure of Peptidoglycan from Thermus thermophilus HB8". JOURNAL OF BACTERIOLOGY, 1995, Vol. 177. p. 4947–4962. http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=177270&blobtype=pdf
2. Henne A, Bruggemann H, Raasch C, Wiezer A, Hartsch T, Liesegang H, Johann A, Lienard T, Gohl O, Martinez-Arias R, Jacobi C, Starkuviene V, Schlenczeck S, Dencker S, Huber R, Klenk HP, Kramer W, Merkl R, Gottschalk G, Fritz HJ. "The genome sequence of the extreme thermophile Thermus thermophilus". Nat Biotechnol. 2004 May, Vol 22. p.547-53. http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=Retrieve&db=PubMed&list_uids=15064768&dopt=AbstractPlus&holding=f1000%2Cf1000m%2Cisrctn
3. Pantazaki AA, Tambaka MG, Langlois V, Guerin P, Kyriakidis DA. "Polyhydroxyalkanoate (PHA) biosynthesis in Thermus thermophilus: purification and biochemical properties of PHA synthase". Mol Cell Biochem. 2003 Dec;254(1-2):173-83. http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=Retrieve&db=PubMed&list_uids=14674696&dopt=AbstractPlus&holding=f1000%2Cf1000m%2Cisrctn
4.Rehse PH, Kitao T, Tahirov TH. "Structure of a closed-form uroporphyrinogen-III C-methyltransferase from Thermus thermophilus". Acta Crystallogr D Biol Crystallogr. 2005 Jul;61(Pt 7):913-9. Epub 2005 Jun 24. http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=Retrieve&db=pubmed&dopt=AbstractPlus&list_uids=15983414&query_hl=1
5. S Takamatsu, R Kato and S Kuramitsu. "Mismatch DNA recognition protein from an extremely thermophilic bacterium, Thermus thermophilus". Nucleic Acids Research, Vol 24, Issue 4 640-647. http://nar.oxfordjournals.org/cgi/content/abstract/24/4/640
6. François J. Franceschi. Max-Planck-Institute for Molecular Genetics http://www.molgen.mpg.de/~ag_ribo/ag_franceschi/franceschi-projects-30S.html
7. Mohamed N, Elfaitouri A, Fohlman J, Friman G, Blomberg J. A sensitive and quantitative single-tube real-time reverse transcriptase-PCR for detection of enteroviral RNA. J Clin Virol. 2004 Jun;30(2):150-6. http://www.ncbi.nlm.nih.gov/sites/entrez?Db=pubmed&Cmd=ShowDetailView&TermToSearch=15125871&ordinalpos=1&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum