Bunyaviridae: Difference between revisions
No edit summary |
No edit summary |
||
| (14 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
{{Curated}} | |||
{{Viral Biorealm Family}} | |||
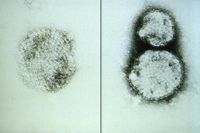
[[Image:6074_lores.jpg|thumb|200px|right|Electron micrograph of the hantavirus responsible for hantavirus pulmonary syndrome, or HPS. From the CDC.]] | |||
==Baltimore Classification== | ==Baltimore Classification== | ||
===Higher order taxa=== | ===Higher order taxa=== | ||
Virus; ssRNA negative-strand viruses; Bunyaviridae | |||
===Genera=== | ===Genera=== | ||
''[[Hantavirus]]'', ''Orthobunyavirus'', ''Nairovirus'', ''Phlebovirus'', ''Tospovirus'' | |||
==Description and Significance== | |||
Bunyaviridae is the largest family of viruses, with over 200 species. | |||
==Genome Structure== | |||
The bunyavirus genome is monomeric and consists of three segments of linear negative-sense (or ambisense, depending on the genus) RNA. The terminal sequences of each segment are base-paired. Because of this, the RNAs form non-covalently closed circles. The nucleotide sequences at the 3'-terminus and the 5'-terminus are complimentary, forming panhandle structures. The 5'-terminus is not capped. | |||
The complete genome is 10500-22700 nucleotides long. The three segments of the genome are labeled L, M, and S. The L segment is 6300-12000 nucleotides long and encodes the viral RNA polymerase. The M segment is 3500-6000 nucleotides long and encodes two glycoproteins as a single gene product that is usually co-translationally cleaved. The S segments is 1000-2200 nucleotides long and encodes the coat protein. (sources: [http://www.dpvweb.net/notes/showfamily.php?family=Bunyaviridae Descriptions of Plant Viruses], [http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/11000000.htm ICTVdB]) | |||
==Virion Structure of a Bunyavirus== | |||
Bunyavirus virions have a complex construction. They consist of an envelope and three nucleocapsids, which each old one segments of the genome. Virions are 80-120nm in diameter. The virions vary in shape from spherical to pleomorphic. Surface projections are 5-10nm long spikes which evenly coat the surface. The nucleocapsids are elongated and have helical symmetry. (source: [http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/11000000.htm ICTVdB]) | |||
==Reproduction Cycle of a Bunyavirus in a Host Cell== | |||
The bunyavirus virion attaches to the surface of a host cell through the G1/G2 glycoproteins and is endocytosed into the host cell. The virus membrane fuses with the vesicle membrane through acidification of the endosome and the conformational change that results from it, and the nucleocapsids are released into the cytoplasm. The RNA-dependent RNA polymerase known as the nucleocapsid-associated L protein interacts with the three S, M, and L nucleocapsids. Primary transcription and translation take place. A switch from translation to replication occurs, and full-length cRNA templates are produced, from which full-length genomes are produced and encapsidated. Virions are assembled and bud in the Golgi. The virions are transported to the cell surface in vesicles and are released from the cell. (source: [http://athena.bioc.uvic.ca/bioDoc/bunyaviridae/ Viral Bioinformatics Resource Center]) | |||
== | ==Viral Ecology & Pathology== | ||
The natural hosts for bunyaviruses are a wide variety of arthropods and mammals. Transmission is usually by parasitic insects (such as ticks and mosquitos) or contact with infected animals or humans. The effects of these viruses on host organisms vary greatly, due to the large size of the family. Generally, once contracted, the virus replicates in specific target organs (which vary depending on the virus, as does severity). | |||
One genus of the bunyaviridae family, the tospoviruses, infect plants. Tospoviruses are transmitted to plants my minute insects called thrips. Tospoviruses (named for the first virus in the genus to be identified, the ''Tomato spotted wilt virus'') cause significant crop losses worldwide. (sources: Weber and Elliott, [http://edis.ifas.ufl.edu/pdffiles/PP/PP13400.pdf Adkins et al.]) | |||
== | ==References== | ||
[http://edis.ifas.ufl.edu/pdffiles/PP/PP13400.pdf Adkins, Scott, Tom Zitter, Tim Momol. Fact Sheet: "Tospoviruses (Family ''Bunyaviridae'' Genus ''Tospovirus'')". October 2005. University of Florida IFAS Extension.] | |||
[http://www.dpvweb.net/notes/showfamily.php?family=Bunyaviridae Descriptions of Plant Viruses: Notes on Family: Bunyaviridae] | |||
[http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/11000000.htm ICTVdB - The Universal Virus Database, version 3. http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/] | |||
[http://athena.bioc.uvic.ca/bioDoc/bunyaviridae/ Viral Bioinformatics Resource Center & Viral Bioinformatics - Canada: Bunyaviridae] | |||
Weber, Friedemann and Richard M. Elliott. "Antigenic drift, antigenic shift, and interferon antagonists: how bunyaviruses counteract the immune system." ''Virus Research'' 88.1-2 (2002): 129-136. | |||
Latest revision as of 00:22, 8 August 2010
A Viral Biorealm page on the family Bunyaviridae
Baltimore Classification
Higher order taxa
Virus; ssRNA negative-strand viruses; Bunyaviridae
Genera
Hantavirus, Orthobunyavirus, Nairovirus, Phlebovirus, Tospovirus
Description and Significance
Bunyaviridae is the largest family of viruses, with over 200 species.
Genome Structure
The bunyavirus genome is monomeric and consists of three segments of linear negative-sense (or ambisense, depending on the genus) RNA. The terminal sequences of each segment are base-paired. Because of this, the RNAs form non-covalently closed circles. The nucleotide sequences at the 3'-terminus and the 5'-terminus are complimentary, forming panhandle structures. The 5'-terminus is not capped.
The complete genome is 10500-22700 nucleotides long. The three segments of the genome are labeled L, M, and S. The L segment is 6300-12000 nucleotides long and encodes the viral RNA polymerase. The M segment is 3500-6000 nucleotides long and encodes two glycoproteins as a single gene product that is usually co-translationally cleaved. The S segments is 1000-2200 nucleotides long and encodes the coat protein. (sources: Descriptions of Plant Viruses, ICTVdB)
Virion Structure of a Bunyavirus
Bunyavirus virions have a complex construction. They consist of an envelope and three nucleocapsids, which each old one segments of the genome. Virions are 80-120nm in diameter. The virions vary in shape from spherical to pleomorphic. Surface projections are 5-10nm long spikes which evenly coat the surface. The nucleocapsids are elongated and have helical symmetry. (source: ICTVdB)
Reproduction Cycle of a Bunyavirus in a Host Cell
The bunyavirus virion attaches to the surface of a host cell through the G1/G2 glycoproteins and is endocytosed into the host cell. The virus membrane fuses with the vesicle membrane through acidification of the endosome and the conformational change that results from it, and the nucleocapsids are released into the cytoplasm. The RNA-dependent RNA polymerase known as the nucleocapsid-associated L protein interacts with the three S, M, and L nucleocapsids. Primary transcription and translation take place. A switch from translation to replication occurs, and full-length cRNA templates are produced, from which full-length genomes are produced and encapsidated. Virions are assembled and bud in the Golgi. The virions are transported to the cell surface in vesicles and are released from the cell. (source: Viral Bioinformatics Resource Center)
Viral Ecology & Pathology
The natural hosts for bunyaviruses are a wide variety of arthropods and mammals. Transmission is usually by parasitic insects (such as ticks and mosquitos) or contact with infected animals or humans. The effects of these viruses on host organisms vary greatly, due to the large size of the family. Generally, once contracted, the virus replicates in specific target organs (which vary depending on the virus, as does severity).
One genus of the bunyaviridae family, the tospoviruses, infect plants. Tospoviruses are transmitted to plants my minute insects called thrips. Tospoviruses (named for the first virus in the genus to be identified, the Tomato spotted wilt virus) cause significant crop losses worldwide. (sources: Weber and Elliott, Adkins et al.)
References
Descriptions of Plant Viruses: Notes on Family: Bunyaviridae
ICTVdB - The Universal Virus Database, version 3. http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/
Viral Bioinformatics Resource Center & Viral Bioinformatics - Canada: Bunyaviridae
Weber, Friedemann and Richard M. Elliott. "Antigenic drift, antigenic shift, and interferon antagonists: how bunyaviruses counteract the immune system." Virus Research 88.1-2 (2002): 129-136.