Host Dependency of Mycobacterium leprae: Difference between revisions
No edit summary |
|||
| (33 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
{{Curated}} | |||
==Introduction== | ==Introduction== | ||
<br> | <br> | ||
[[Image:M._leprae_in_human_tissue.jpg|thumb|200px|right|''Mycobacterium leprae in human tissue, photo taken from presentation by Chow E and Roman T | [[Image:M._leprae_in_human_tissue.jpg|thumb|200px|right|''<i>Mycobacterium leprae</i> in human tissue, photo taken from presentation by Chow E and Roman T]] | ||
Mycobacterium leprae, the bacterial cause of leprosy, is almost impossible to culture in a laboratory. M. leprae is an acid fast gram positive bacillus, | <i>Mycobacterium leprae</i>, the bacterial cause of leprosy, is almost impossible to culture in a laboratory (Slonczewski, 2009). <i>M. leprae</i> is an acid fast gram positive bacillus (Slonczewski, 2009). <i>M. leprae</i> has one of the slowest doubling times of any pathogen. It takes approximately 14 days for the cells to divide (Slonczewski, 2009). <i>M. leprae</i> is easily detected on Fite-Faraco staining (Ooi,2004). <i>M. leprae</i> is from the same genus as <i> Mycobacterium tuberculosis</i>; the two species have similar physical characteristics and similar genomes. | ||
The genome for M. leprae has been sequenced, and it has been found that almost half of the genes are pseudogenes; genes that no longer code for proteins to be transcribed in the cell. Many of these pseudogenes correspond to genes found in Mycobacterium tuberculosis that are still functional ( | The genome for M. leprae has been sequenced, and it has been found that almost half of the genes are pseudogenes; genes that no longer code for proteins to be transcribed in the cell (Cole, 2001). Many of these pseudogenes correspond to genes found in Mycobacterium tuberculosis that are still functional (Cole, 2001). The loss of these genes have caused <i>M. leprae</i> to rely on the host cell to survive. The bacteria needs an extremely specific environment to thrive in. | ||
<i>M. leprae</i> has plagued mankind since ancient times. Antibiotics have helped to decrease the number of cases of leprosy, but poorer areas around the world still have problems with this disease. In 2004, there were approximately 50,000 new cases of infection. | It is extremely difficult to culture <i>Mycobacterium leprae</i>. All attempts to create a medium that the bacteria are able to grow in has failed. Scientists have found that the bacteria can only grow when acting as a parasite in animals with lower body temperature, such as armadillos, genetically immune deficient mice, or the extremities of a human body (Slonczewski 2009). Aramadillos are used predominantly as a host for the bacteria, but the animals are difficult to work with, thus making animal research on <i>Mycobacterium leprae</i> slow and complicated (Wheeler, 2002). Scientists are still attempting to create a media that will support growth of the bacteria to increase the ease of studying it. | ||
<i>M. leprae</i> has plagued mankind since ancient times. Leprosy was one of the most terrifying diseases in history due to the large open sores and deformations it caused. Hundreds of thousands of people died each year from leprosy until drugs were found that could combat it. Antibiotics have helped to decrease the number of cases of leprosy, but poorer areas around the world still have problems with this disease. In 2004, there were approximately 50,000 new cases of infection. | |||
<br> | <br> | ||
==Mycobacterium leprae genome; missing genes in M. leprae compared to M. tuberculosis== | ==<i>Mycobacterium leprae</i> genome; missing genes in <i>M. leprae</i> compared to <i>M. tuberculosis</i>== | ||
<br> | <br> | ||
The <i>Mycobacterium leprae</i> genome was sequenced by S. T. Cole and was published in Nature (Cole 2001). His group found that the genome consisted of a single circular chromosome approximately 3,268,203 bp. The G-C content of the genome is 57.7%, and only 49.5% of the genome still codes for proteins (Cole 2001). The genomes for <i>M. leprae</i> and <i>M. tuberculosis</i> are quite similar, and about 27% of <i>M. leprae</i>'s genome are pseudogenes that directly correspond to genes still expressed in <i>M. tuberculosis</i> (Cole 2001). The remaining 23% of the genome does not resemble coding genes; these sections seem to have mutated over time and are currently unrecognizable. The similarity of the pseudogenes in <i>M. leprae</i> compared to expressed genes in <i>M. tuberculosis</i> suggest that the genome of <i>M. leprae</i> has evolved to lose these genes because they were not necessary for survival. This is an example of reductive evolution. It is estimated that <i>M. leprae</i> has lost approximately 2000 genes from its genome (Cole 2001). The loss of genes in metabolic pathways such as energy metabolism, limiting the carbon sources <i>M leprae</i> can use, and holes in respiration pathways help to explain why <i>M leprae</i> can not be cultured in a laboratory and has only been shown to infect humans, the footpads of mice, and armadillos. | |||
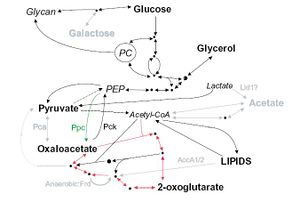
[[Image:Carbon_Metabolism_in_M_leprae_jpeg.JPG|thumb|500 x 500px||right|Figure 1. Carbon Metabolism in <i>M. leprae</i> compared to <i>M. tuberculosis</i>. Pathways in black are in both <i>M. leprae</i> and <i>M. tuberculosis</i>, grey pathways are pathways that exist in <i>M. tuberculosis</i> that <i>M. leprae</i> still has pseudogenes for. Bolded carbon sources are possible for use by <i>M. leprae</i>, grey carbon sources are unusable by <i>M leprae</i>. The pathway with the red arrows is the Krebs cycle. The figure was taken from Wheeler 2002.]] | |||
From a thorough examination of the genome of <i>M. leprae</i>, it was found that <i>M leprae</i> does have all the main metabolic and biosynthetic pathways, but a there are some missing genes (Wheeler, 2002). Compared to the genome of <i>M. tuberculosis</i>, <i>M leprae</i> lacks the genes for vitamin B12 synthesis and metC, a gene that codes for the enzyme that converts cysteine into methionine (Wheeler, 2002). This means that <i>M leprae</i> can not synthesize the vitamin or methionine, an amino acid, and instead has to be taken from the environment (Wheeler 2002). To test the presence of methionine and vitamin B12 were enough for <i>M leprae</i> to grow, an attempt to culture the bacteria in media containing these molecules has not successful. | |||
There are significant losses in the genome of <i>M. leprae</i> for carbon and energy metabolism. All the central pathways are present; <i>M. leprae</i> metabolizes glucose, but other carbon sources can not be metabolized for energy. There are pseudogenes present in <i>M. leprae</i> corresponding to genes in <i>M. tuberculosis</i> for proteins that enable the bacteria to breakdown other carbon sources such as acetate and galactose (wheeler 2002). Figure 1 shows carbon metabolism in <i>M. leprae</i> compared to <i>M. tuberculosis</i>. Pathways in black are present in both <i>M. leprae</i> and <i>M. tuberculosis</i>, grey pathways are pathways that exist in <i>M. tuberculosis</i> that <i>M. leprae</i> still has pseudogenes for. Bolded carbon sources are possible for use by <i>M. leprae</i>, grey carbon sources are unusable by <i>M. leprae</i>. The pathway with the red arrows is the Krebs cycle. Not being able to use these sources for energy means that <i>M. leprae</i> has to rely much more heavily on using glucose, which is not found in the environment as commonly as other carbon sources (Wheeler 2002). Most microbes are able to convert other sources such as galactose into glucose, but <i>M. leprae</i> is not able to do this and must rely on another organism to make sure it has enough glucose. | |||
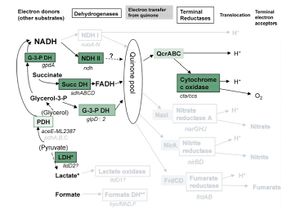
[[Image:Respiration_in_M_leprae.JPG|thumb|500 x 500px||left|Figure 2. Respiration in M. leprae compared to M. tuberculosis. Pathways in black are in both M. leprae and M. tuberculosis, grey pathways are pathways that exist in M. tuberculosis that M. leprae still has pseudogenes for. The figure was taken from Wheeler 2002.]] | |||
<i>M. leprae</i> has the genes for the proteins for the Krebs' cycle and glyoxylate cycle, but the organism essentially relies on a single electron donor, FADH. | <i>M. leprae</i> has the genes for the proteins for the Krebs' cycle and glyoxylate cycle, but the organism essentially relies on a single electron donor, FADH (Wheeler 2002). Figure 2 shows respiration pathways for <i>M. leprae</i> compared to <i>M. tuberculosis</i>. As in the previous figure, pathways in black are present in both <i>M. leprae</i> and <i>M. tuberculosis</i>, grey pathways are pathways that exist in <i>M. tuberculosis</i> that <i>M. leprae</i> still has pseudogenes for. NADH, one of the more common electron donors involved in metabolism in many species, is not used for electron transport in <i>M. leprae</i>. This causes aerobic respiration to be much less efficient. There are no anaerobic respiration pathways in <i>M. leprae</i>, so the bacteria have to make due with a less efficient aerobic pathway (Wheeler 2002). This makes the bacteria even harder to culture due to the difficulty of composing a media with the correct levels of oxygen and the other necessary vitamins and minerals to permit growth. <i>M. leprae</i> has evolved to only survive in it's niche environment with a host. | ||
| Line 43: | Line 43: | ||
<br> | <br> | ||
It has been shown that <i>M. leprae</i> are able to bind to nasal epithelial cells by binding to a soluble protein, fibronectin, that binds to fibronectin receptors on the surface of the epithelial cell | <i>Mycobacterium leprae</i> primarily infects the lower temperature extremities, such as the epithelial cells and nonmyelin producing Schwann cells around peripheral nerves in the hands and feet, and occasionally the upper respiratory tract, testes, and cornea, causing the disease leprosy. The symptoms of leprosy include anesthetic skin lesions and enlarged peripheral nerves. Infected areas witness less sensation to pain and temperature. Infection usually follows breathing in through the nose microscopic droplets excreted from an infected individual containing <i>M. leprae</i>, usually from sneezing. There are also cases of infection when a human comes into contact with soil containing <i>M. leprae</i>. Skin contact with an infected individual has not proven to pass on the infection (Ooi, 2004). The number of cases of individuals infected with leprosy have decreased overtime with the help of antibiotics, but there are still hundreds of thousands of cases of leprosy across the world. | ||
It has been shown that <i>M. leprae</i> are able to bind to nasal epithelial cells by binding to a soluble protein, fibronectin, that binds to fibronectin receptors on the surface of the epithelial cell (Byrd 1993). It is thought that <i>M. leprae</i> then enter the nasal epithelial cells, enter the blood stream, and migrate to places with the best environment, the nonmyelinating Schwann cells in the extremities (Brophy 2002). It was shown that <i>M. leprae</i> invade the nonmyelinating Schwann cells and multiply, as well as attach to myelinating Schwann cells (Brophy 2002). The <i>M. leprae</i> cells release PGL proteins, which are thought to disrupt the DRP2-dystroglycan complex in the Schwann cell, causing the myelination to deteriorate (Brophy 2002). Myelinating Schwann cells produce DRP2, which is necessary for myelination. The dystroglycan complexes link the Schwann cells and are thought to carry signals from the inside of the cell to the outside (Ooi 2004). It has been shown that dead <i>M. leprae</i> cells or the cell wall alone can still cause demyelination to occur (Brophy 2002). This means that, even after antibiotics are administered and the bacteria are killed, more demyelination occurs. This is why it is important that the infection is treated right away; before too much damage to the nervous system is done. | |||
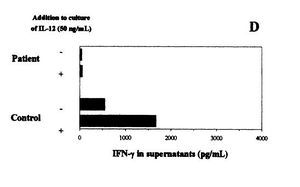
[[Image:Leprae_figure.JPG|thumb|400x 400px||right|Figure 3. Effects of IL-12 in IFN-y production in individuals infected with <i>M. leprae</i> and healthy individuals. IL-12 caused production of IFN-y to increase in the healthy individual, but not in the infected individual. Figure from Delobel, 2003.]] | |||
<i>M. leprae</i> have been also noted to cause inflammation in the extremities of infected individuals by turning off the immune system. On the cell wall of <i>M. leprae</i> there are lipids that suppress T-cell activation without altering the membrane of the T cells (Moura 1997). By inhibiting the immune system, <i>M. leprae</i> is able continue to thrive without being attacked. Figure 3 shows the results of an experiment comparing the levels of IFN-y product produced by a patient infected with <i>M. leprae</i> and a healthy individual (Delobel, 2003). IFN-y are interferons produced by T cells and are one of the body's responses to fight a microbial infection (Ooi, 2004). The levels of IFN-y product were recorded over a 72 hour time frame, as shown in the figure as the '-' for each person. The + corresponds to the levels of IFN-y product 72 hours after administering IL-12, a protein that regulates the production of IFN-y by TH1 helper cells. It is clear from these results that the IL-12 increased the levels of IFN-y from the healthy individual, but not had no effect on the PBMC of the individual infected with <i>M. leprae</i> (Delobel, 2003). The bacteria stopped the immune system from increasing the production of IFN-y that is harmful to the bacteria when IL-12 was administered. <i>M. leprae</i> successfully suppressed the immune response fight off the infection. | |||
It is because of <i>M. leprae</i>'s ability to disarm the immune system that the infection is extremely hard to get rid of naturally. Before the time of antibiotics, the majority of the individuals that became infected and showed symptoms of the disease usually died from it. | |||
==Genetic component of the host== | ==Genetic component of the host== | ||
<br> | <br> | ||
Depending on the host, the severity of an infection of <i>Mycobacterium leprae</i> can range from no infection to an extreme case of leprosy. How well the bacteria grow in the body depends on the genetics and the state of the immune system of the host. Some individuals are more susceptible than others, for genetic reasons. Leprosy susceptibility has been shown to be determined by genetics by family and twin studies.Over 90% of individuals infected with develop immunity without showing any symptoms (Ooi 2004). Those that are susceptible to the disease do not necessarily have a general immunity deficiency. There are three main forms of leprosy from one side of the spectrum to the other that are commonly found, as described in table 3 (Ooi, 2004). This is known as the Ridley-Joping classification of the disease. The spectrum shows the range of individuals that have T helper 1 (TH1) and T helper 2 (TH2) immune responses capable of killing <i>M. leprae</i> results in no infection, while those that are incapable of destroying the bacteria develop the more extreme lepromatous leprosy. | |||
[[Image:different_types_of_leprosy.JPG|thumb|500x 400px||left|Table 3-Different types of leprosy and the symptoms/severity of each type compared to eachother. The figure was taken from Ooi, W 2004.]] | |||
The T cells in the immune system are the foundation of the cell mediated immunity. Two types of T cells fight infection. It is the TH2 cells, the T helper cells that are used to take detect bacteria growing outside of cells in the body. T cells have receptors on the outside surface of the cells that are used to detect a wide variety of antigens such as invading species like <i>Mycobacterium leprae</i>. When the T helper cells detect the bacteria, the T cells activate B cells, the part of the immune system that creates antibodies, to create antibodies to cause the bacteria to be destroyed by phagocytes. The TH1 cells respond to antigens on host cells infected with the bacteria. The TH1 cells then activate Tc cells to destroy the infected cells by releasing cytokines into the cell. | |||
The protein NRAMP1, HLA genes, TLR1 and TLR2 receptors, and PARK2 and PACRG genes all influence which form of the infection will develop, if an infection occurs (Ooi, 2004, Schurr, 2007). Each gene or protein subtly changes the way either the T cells or host cells interact with <i>Mycobacterium leprae</i>, which changes the environment of the bacteria. NRAMP1, the natural resistance associated macrophage protein 1, helps in regulating the development of immune responses targeting mycobacteria (Ooi 2004). The protein helps the immune system fight off the infection by producing more specific microbial antibodies to detect the bacteria in the bloodstream. The effectiveness of the protein at killing <i>Mycobacterium leprae</i> has been linked to specific racial groups (Ooi 2004). Individuals with a normal functioning NRAMP1 are less susceptable to an infection by <i>Mycobacterium leprae</i> because it can be detected in the body faster. (Ooi 2004) HLA genes help determine how effective the immune system is at fighting the infection. Different versions of this gene will cause differences in the type of leprosy the individual contracts (Ooi 2004). The TLR1 and TLR2 toll like receptors determine the level of cytokine secretion during an immunity response. Differences in the genetics of these receptors will change the effectiveness of secreting cytokines, and will have an impact on which type of leprosy the individual comes down with. The presence of PARK2 and PACRG mutations, which occur as a result of a single nucleotide mutation located on chromosome 6q25 have been shown to increase susceptibility to leprosy infections (Schurr 2007). This increased susceptibility has been shown in multiple Vietnamese families. | |||
==Conclusion== | ==Conclusion== | ||
<br> | <br> <i>Mycobacterium leprae</i> can only survive in extremely selective environments as a result of reductive evolution. The bacteria still infect a hundreds of thousands of people a year worldwide, but with the help of antibiotics the number has decreased. New antibiotic resistant strains are appearing, and stronger cocktails of drugs containing multiple antibiotics are now used to treat the disease. The multidrug cocktail currently used consists of the antibiotics Dapsone, rifampin, and clofazimine. It is possible that if the correct combination of minerals, vitamins, and amino acids is found that when added to a media is the correct environment for <i>Mycobacterium leprae</i> it could allow us to study the bacteria more and make drug research more effective. | ||
Even though 90% of those that come into contact with <i>Mycobacterium leprae</i> do not develop an infection, those that do develop an infection show a wide spectrum of severity of symptoms as a result of a few key genes. If an infection is not caught early, there will be residual damage done to the peripheral nervous system of the infected individual because the lipids on the cell wall of the bacteria cause de-myelination of Schwann cells. Donations are needed to help develop cheaper, more effective drugs to kill the bacteria. If you would like to learn more about the fight against leprosy or to donate to help supply drugs to infected individuals that can not afford to purchase the medication, go to the website of the organization Lepra at http://www.lepra.org.uk/home.asp. | |||
==References== | ==References== | ||
Ascenzi, P., Bolognesi, M., Visca, P. NO Dissociation Represents the Rate Limiting step for O2-Mediated Oxidation of Ferrous Nitrosylated <i>Mycobacterium leprae</i> truncated hemoglobin O. <i>BBRC</i>, 357. 2007. 809-814. | |||
Brophy, P. Subversion of Schwann cells and the leper's bell. <i>Science</i> 296. 2002. 862-863 | |||
Byrd, S., Gelber, R., Bermudez, L. Roles of Soluble Fibronectin and B1 Integrin Receptors in the Binding of <i>Mycobacterium leprae</i> to Nasal Epithelial Cells. <i>Clinical Immunology and Ummunopathology</i> Vol 69, no.3 .1993. 266-271 | |||
Cole, S. Massive gene decay in the leprosy bacillus. <i>Nature</i> 409 2001. 1007-1011 | |||
Delobel, P. Launois, P., Djossou, F. Sainte-Marie D., Pradinaud, R. American Cutanerous Leishmaniasis, Lepromatous Leprosy, Pulmonary Tuberculosis Coinfection with Downregulation of the T Helper Cell Response. <i>Clinical Infectious Diseases</i>, Vol 37, 5. 2003. pp 628-633 | |||
Geluk, A., Ottenhoff, T. HLA and Leprosy in the Pre and Postgenomic Eras. <i>Human Immunology</i>, 67. 2006. 439-445. | |||
Moura, A., Mariano, M. Lipids from <i>Mycobacterium leprae</i> cell wall suppress T-cell activation in vivo and in vitro. <i>Immunology</i>, 92. 1997. 429-436. | |||
Ooi, W., Srinivasan, J. Leprosy and the Peripheral Nervous System: Basic and Clinical Aspects. <i>Muscle and Nerve</i>, 2004. 393-409. | |||
Schurr, E., Alcais, A., Singh M., Mehra, N., Abel, L. Mycobacterial infections: PARK2 and PACRG associations in Leprosy. <i>Tissue Antigens</i>, 2007. 231-233. | |||
Slonczewski, J., Foster, J. <b>Microbiology: An Evolving Science</b>. New York: W. W. Norton, 2009. 700-702. | |||
Wiker, H., Spierings, E., Kolkman, M, Ottenhoff, T., Harboe, M. The mammalian cell entrypoeron 1 (mce1) of <i>Mycobacterium Leprae</i> and <i>Mycobacterium tuberculosis</i>. <i>Microbial Pathogenesis</i>, 27. 1999. 173-177. | |||
Wheeler, P. Leprosy- Clues about the Biochemistry of <i>Mycobacterium leprae</i> and its Host-Dependency from the Genome. <i>World Journal of Microbiology and Biochemistry</i>, 19. 2003. 1-16. | |||
http://images.google.com/imgres?imgurl=http://www.stanford.edu/group/parasites/ParaSites2008/Eric%2520Chow_Tania%2520Roman/Leprosy_files/image002.jpg&imgrefurl=http://www.stanford.edu/group/parasites/ParaSites2008/Eric%2520Chow_Tania%2520Roman/Leprosy.htm&usg=__FuMa0EbNt90SLHIsPNtHoPG_L-U=&h=282&w=415&sz=37&hl=en&start=12&sig2=QhgkDewUDwUHwVGbYZ7zJQ&tbnid=SluTAPufmlK5lM:&tbnh=85&tbnw=125&prev=/images%3Fq%3Dmycobacterium%2Bleprae%26gbv%3D2%26hl%3Den%26sa%3DG&ei=sR7mSd6pK5T0nQfQ4v2XDg'' | |||
Edited by student of [mailto:slonczewski@kenyon.edu Joan Slonczewski] for [http://biology.kenyon.edu/courses/biol238/biol238syl09.html BIOL 238 Microbiology], 2009, [http://www.kenyon.edu/index.xml Kenyon College]. | Edited by student of [mailto:slonczewski@kenyon.edu Joan Slonczewski] for [http://biology.kenyon.edu/courses/biol238/biol238syl09.html BIOL 238 Microbiology], 2009, [http://www.kenyon.edu/index.xml Kenyon College]. | ||
Latest revision as of 20:14, 10 August 2010
Introduction
Mycobacterium leprae, the bacterial cause of leprosy, is almost impossible to culture in a laboratory (Slonczewski, 2009). M. leprae is an acid fast gram positive bacillus (Slonczewski, 2009). M. leprae has one of the slowest doubling times of any pathogen. It takes approximately 14 days for the cells to divide (Slonczewski, 2009). M. leprae is easily detected on Fite-Faraco staining (Ooi,2004). M. leprae is from the same genus as Mycobacterium tuberculosis; the two species have similar physical characteristics and similar genomes.
The genome for M. leprae has been sequenced, and it has been found that almost half of the genes are pseudogenes; genes that no longer code for proteins to be transcribed in the cell (Cole, 2001). Many of these pseudogenes correspond to genes found in Mycobacterium tuberculosis that are still functional (Cole, 2001). The loss of these genes have caused M. leprae to rely on the host cell to survive. The bacteria needs an extremely specific environment to thrive in.
It is extremely difficult to culture Mycobacterium leprae. All attempts to create a medium that the bacteria are able to grow in has failed. Scientists have found that the bacteria can only grow when acting as a parasite in animals with lower body temperature, such as armadillos, genetically immune deficient mice, or the extremities of a human body (Slonczewski 2009). Aramadillos are used predominantly as a host for the bacteria, but the animals are difficult to work with, thus making animal research on Mycobacterium leprae slow and complicated (Wheeler, 2002). Scientists are still attempting to create a media that will support growth of the bacteria to increase the ease of studying it.
M. leprae has plagued mankind since ancient times. Leprosy was one of the most terrifying diseases in history due to the large open sores and deformations it caused. Hundreds of thousands of people died each year from leprosy until drugs were found that could combat it. Antibiotics have helped to decrease the number of cases of leprosy, but poorer areas around the world still have problems with this disease. In 2004, there were approximately 50,000 new cases of infection.
Mycobacterium leprae genome; missing genes in M. leprae compared to M. tuberculosis
The Mycobacterium leprae genome was sequenced by S. T. Cole and was published in Nature (Cole 2001). His group found that the genome consisted of a single circular chromosome approximately 3,268,203 bp. The G-C content of the genome is 57.7%, and only 49.5% of the genome still codes for proteins (Cole 2001). The genomes for M. leprae and M. tuberculosis are quite similar, and about 27% of M. leprae's genome are pseudogenes that directly correspond to genes still expressed in M. tuberculosis (Cole 2001). The remaining 23% of the genome does not resemble coding genes; these sections seem to have mutated over time and are currently unrecognizable. The similarity of the pseudogenes in M. leprae compared to expressed genes in M. tuberculosis suggest that the genome of M. leprae has evolved to lose these genes because they were not necessary for survival. This is an example of reductive evolution. It is estimated that M. leprae has lost approximately 2000 genes from its genome (Cole 2001). The loss of genes in metabolic pathways such as energy metabolism, limiting the carbon sources M leprae can use, and holes in respiration pathways help to explain why M leprae can not be cultured in a laboratory and has only been shown to infect humans, the footpads of mice, and armadillos.
From a thorough examination of the genome of M. leprae, it was found that M leprae does have all the main metabolic and biosynthetic pathways, but a there are some missing genes (Wheeler, 2002). Compared to the genome of M. tuberculosis, M leprae lacks the genes for vitamin B12 synthesis and metC, a gene that codes for the enzyme that converts cysteine into methionine (Wheeler, 2002). This means that M leprae can not synthesize the vitamin or methionine, an amino acid, and instead has to be taken from the environment (Wheeler 2002). To test the presence of methionine and vitamin B12 were enough for M leprae to grow, an attempt to culture the bacteria in media containing these molecules has not successful.
There are significant losses in the genome of M. leprae for carbon and energy metabolism. All the central pathways are present; M. leprae metabolizes glucose, but other carbon sources can not be metabolized for energy. There are pseudogenes present in M. leprae corresponding to genes in M. tuberculosis for proteins that enable the bacteria to breakdown other carbon sources such as acetate and galactose (wheeler 2002). Figure 1 shows carbon metabolism in M. leprae compared to M. tuberculosis. Pathways in black are present in both M. leprae and M. tuberculosis, grey pathways are pathways that exist in M. tuberculosis that M. leprae still has pseudogenes for. Bolded carbon sources are possible for use by M. leprae, grey carbon sources are unusable by M. leprae. The pathway with the red arrows is the Krebs cycle. Not being able to use these sources for energy means that M. leprae has to rely much more heavily on using glucose, which is not found in the environment as commonly as other carbon sources (Wheeler 2002). Most microbes are able to convert other sources such as galactose into glucose, but M. leprae is not able to do this and must rely on another organism to make sure it has enough glucose.
M. leprae has the genes for the proteins for the Krebs' cycle and glyoxylate cycle, but the organism essentially relies on a single electron donor, FADH (Wheeler 2002). Figure 2 shows respiration pathways for M. leprae compared to M. tuberculosis. As in the previous figure, pathways in black are present in both M. leprae and M. tuberculosis, grey pathways are pathways that exist in M. tuberculosis that M. leprae still has pseudogenes for. NADH, one of the more common electron donors involved in metabolism in many species, is not used for electron transport in M. leprae. This causes aerobic respiration to be much less efficient. There are no anaerobic respiration pathways in M. leprae, so the bacteria have to make due with a less efficient aerobic pathway (Wheeler 2002). This makes the bacteria even harder to culture due to the difficulty of composing a media with the correct levels of oxygen and the other necessary vitamins and minerals to permit growth. M. leprae has evolved to only survive in it's niche environment with a host.
Interaction with host cells
Mycobacterium leprae primarily infects the lower temperature extremities, such as the epithelial cells and nonmyelin producing Schwann cells around peripheral nerves in the hands and feet, and occasionally the upper respiratory tract, testes, and cornea, causing the disease leprosy. The symptoms of leprosy include anesthetic skin lesions and enlarged peripheral nerves. Infected areas witness less sensation to pain and temperature. Infection usually follows breathing in through the nose microscopic droplets excreted from an infected individual containing M. leprae, usually from sneezing. There are also cases of infection when a human comes into contact with soil containing M. leprae. Skin contact with an infected individual has not proven to pass on the infection (Ooi, 2004). The number of cases of individuals infected with leprosy have decreased overtime with the help of antibiotics, but there are still hundreds of thousands of cases of leprosy across the world.
It has been shown that M. leprae are able to bind to nasal epithelial cells by binding to a soluble protein, fibronectin, that binds to fibronectin receptors on the surface of the epithelial cell (Byrd 1993). It is thought that M. leprae then enter the nasal epithelial cells, enter the blood stream, and migrate to places with the best environment, the nonmyelinating Schwann cells in the extremities (Brophy 2002). It was shown that M. leprae invade the nonmyelinating Schwann cells and multiply, as well as attach to myelinating Schwann cells (Brophy 2002). The M. leprae cells release PGL proteins, which are thought to disrupt the DRP2-dystroglycan complex in the Schwann cell, causing the myelination to deteriorate (Brophy 2002). Myelinating Schwann cells produce DRP2, which is necessary for myelination. The dystroglycan complexes link the Schwann cells and are thought to carry signals from the inside of the cell to the outside (Ooi 2004). It has been shown that dead M. leprae cells or the cell wall alone can still cause demyelination to occur (Brophy 2002). This means that, even after antibiotics are administered and the bacteria are killed, more demyelination occurs. This is why it is important that the infection is treated right away; before too much damage to the nervous system is done.
M. leprae have been also noted to cause inflammation in the extremities of infected individuals by turning off the immune system. On the cell wall of M. leprae there are lipids that suppress T-cell activation without altering the membrane of the T cells (Moura 1997). By inhibiting the immune system, M. leprae is able continue to thrive without being attacked. Figure 3 shows the results of an experiment comparing the levels of IFN-y product produced by a patient infected with M. leprae and a healthy individual (Delobel, 2003). IFN-y are interferons produced by T cells and are one of the body's responses to fight a microbial infection (Ooi, 2004). The levels of IFN-y product were recorded over a 72 hour time frame, as shown in the figure as the '-' for each person. The + corresponds to the levels of IFN-y product 72 hours after administering IL-12, a protein that regulates the production of IFN-y by TH1 helper cells. It is clear from these results that the IL-12 increased the levels of IFN-y from the healthy individual, but not had no effect on the PBMC of the individual infected with M. leprae (Delobel, 2003). The bacteria stopped the immune system from increasing the production of IFN-y that is harmful to the bacteria when IL-12 was administered. M. leprae successfully suppressed the immune response fight off the infection.
It is because of M. leprae's ability to disarm the immune system that the infection is extremely hard to get rid of naturally. Before the time of antibiotics, the majority of the individuals that became infected and showed symptoms of the disease usually died from it.
Genetic component of the host
Depending on the host, the severity of an infection of Mycobacterium leprae can range from no infection to an extreme case of leprosy. How well the bacteria grow in the body depends on the genetics and the state of the immune system of the host. Some individuals are more susceptible than others, for genetic reasons. Leprosy susceptibility has been shown to be determined by genetics by family and twin studies.Over 90% of individuals infected with develop immunity without showing any symptoms (Ooi 2004). Those that are susceptible to the disease do not necessarily have a general immunity deficiency. There are three main forms of leprosy from one side of the spectrum to the other that are commonly found, as described in table 3 (Ooi, 2004). This is known as the Ridley-Joping classification of the disease. The spectrum shows the range of individuals that have T helper 1 (TH1) and T helper 2 (TH2) immune responses capable of killing M. leprae results in no infection, while those that are incapable of destroying the bacteria develop the more extreme lepromatous leprosy.
The T cells in the immune system are the foundation of the cell mediated immunity. Two types of T cells fight infection. It is the TH2 cells, the T helper cells that are used to take detect bacteria growing outside of cells in the body. T cells have receptors on the outside surface of the cells that are used to detect a wide variety of antigens such as invading species like Mycobacterium leprae. When the T helper cells detect the bacteria, the T cells activate B cells, the part of the immune system that creates antibodies, to create antibodies to cause the bacteria to be destroyed by phagocytes. The TH1 cells respond to antigens on host cells infected with the bacteria. The TH1 cells then activate Tc cells to destroy the infected cells by releasing cytokines into the cell.
The protein NRAMP1, HLA genes, TLR1 and TLR2 receptors, and PARK2 and PACRG genes all influence which form of the infection will develop, if an infection occurs (Ooi, 2004, Schurr, 2007). Each gene or protein subtly changes the way either the T cells or host cells interact with Mycobacterium leprae, which changes the environment of the bacteria. NRAMP1, the natural resistance associated macrophage protein 1, helps in regulating the development of immune responses targeting mycobacteria (Ooi 2004). The protein helps the immune system fight off the infection by producing more specific microbial antibodies to detect the bacteria in the bloodstream. The effectiveness of the protein at killing Mycobacterium leprae has been linked to specific racial groups (Ooi 2004). Individuals with a normal functioning NRAMP1 are less susceptable to an infection by Mycobacterium leprae because it can be detected in the body faster. (Ooi 2004) HLA genes help determine how effective the immune system is at fighting the infection. Different versions of this gene will cause differences in the type of leprosy the individual contracts (Ooi 2004). The TLR1 and TLR2 toll like receptors determine the level of cytokine secretion during an immunity response. Differences in the genetics of these receptors will change the effectiveness of secreting cytokines, and will have an impact on which type of leprosy the individual comes down with. The presence of PARK2 and PACRG mutations, which occur as a result of a single nucleotide mutation located on chromosome 6q25 have been shown to increase susceptibility to leprosy infections (Schurr 2007). This increased susceptibility has been shown in multiple Vietnamese families.
Conclusion
Mycobacterium leprae can only survive in extremely selective environments as a result of reductive evolution. The bacteria still infect a hundreds of thousands of people a year worldwide, but with the help of antibiotics the number has decreased. New antibiotic resistant strains are appearing, and stronger cocktails of drugs containing multiple antibiotics are now used to treat the disease. The multidrug cocktail currently used consists of the antibiotics Dapsone, rifampin, and clofazimine. It is possible that if the correct combination of minerals, vitamins, and amino acids is found that when added to a media is the correct environment for Mycobacterium leprae it could allow us to study the bacteria more and make drug research more effective.
Even though 90% of those that come into contact with Mycobacterium leprae do not develop an infection, those that do develop an infection show a wide spectrum of severity of symptoms as a result of a few key genes. If an infection is not caught early, there will be residual damage done to the peripheral nervous system of the infected individual because the lipids on the cell wall of the bacteria cause de-myelination of Schwann cells. Donations are needed to help develop cheaper, more effective drugs to kill the bacteria. If you would like to learn more about the fight against leprosy or to donate to help supply drugs to infected individuals that can not afford to purchase the medication, go to the website of the organization Lepra at http://www.lepra.org.uk/home.asp.
References
Ascenzi, P., Bolognesi, M., Visca, P. NO Dissociation Represents the Rate Limiting step for O2-Mediated Oxidation of Ferrous Nitrosylated Mycobacterium leprae truncated hemoglobin O. BBRC, 357. 2007. 809-814.
Brophy, P. Subversion of Schwann cells and the leper's bell. Science 296. 2002. 862-863
Byrd, S., Gelber, R., Bermudez, L. Roles of Soluble Fibronectin and B1 Integrin Receptors in the Binding of Mycobacterium leprae to Nasal Epithelial Cells. Clinical Immunology and Ummunopathology Vol 69, no.3 .1993. 266-271 Cole, S. Massive gene decay in the leprosy bacillus. Nature 409 2001. 1007-1011
Delobel, P. Launois, P., Djossou, F. Sainte-Marie D., Pradinaud, R. American Cutanerous Leishmaniasis, Lepromatous Leprosy, Pulmonary Tuberculosis Coinfection with Downregulation of the T Helper Cell Response. Clinical Infectious Diseases, Vol 37, 5. 2003. pp 628-633
Geluk, A., Ottenhoff, T. HLA and Leprosy in the Pre and Postgenomic Eras. Human Immunology, 67. 2006. 439-445.
Moura, A., Mariano, M. Lipids from Mycobacterium leprae cell wall suppress T-cell activation in vivo and in vitro. Immunology, 92. 1997. 429-436.
Ooi, W., Srinivasan, J. Leprosy and the Peripheral Nervous System: Basic and Clinical Aspects. Muscle and Nerve, 2004. 393-409.
Schurr, E., Alcais, A., Singh M., Mehra, N., Abel, L. Mycobacterial infections: PARK2 and PACRG associations in Leprosy. Tissue Antigens, 2007. 231-233.
Slonczewski, J., Foster, J. Microbiology: An Evolving Science. New York: W. W. Norton, 2009. 700-702.
Wiker, H., Spierings, E., Kolkman, M, Ottenhoff, T., Harboe, M. The mammalian cell entrypoeron 1 (mce1) of Mycobacterium Leprae and Mycobacterium tuberculosis. Microbial Pathogenesis, 27. 1999. 173-177.
Wheeler, P. Leprosy- Clues about the Biochemistry of Mycobacterium leprae and its Host-Dependency from the Genome. World Journal of Microbiology and Biochemistry, 19. 2003. 1-16.
Edited by student of Joan Slonczewski for BIOL 238 Microbiology, 2009, Kenyon College.