Potential Therapeutics Isolated from Salinispora: Difference between revisions
No edit summary |
|||
| (42 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{{Curated}} | |||
<br>Christina Kucher<br> | |||
==Introduction== | ==Introduction== | ||
<br>Rather than designing synthetic drugs, some researchers are exploring natural sources for cancer treatments and as models for future synthetic treatments. One promising source includes the recently discovered <i>Salinispora</i> bacteria, belonging to the order <i>Actinomycetales</i> (also known as actinomycetes). This order includes many species that have contributed to today’s naturally occurring antibiotics. It was originally believed that the gram positive <i>Actinomycetales</i> only existed in soil habitats, and that any actinomycetes found in marine environments came from spores produced by terrestrial species (1). However, in search of new actinomycetes as possible dug sources, Jensen and Fenical of the Scripps Institution of Oceanography in the late 1980s discovered actinomycetes (originally called MAR1) that required seawater for growth (2). In 2002, the separate strains of marine origin were found in tropical and subtropical sediment, and classified under the genus <i>Salinispora</i>, belonging to the family <i>Micromonosporaceae</i>. Two species have been formally identified as <i>Salinispora arenicola</i> and <i>Salinispora tropica</i>, while a third species of <i>Salinispora pacifica</i> is being studied (2,3). The <i>Salinispora</i> bacteria have, like the rest of their order, a variety of secondary metabolites that may be useful in the pharmeceutical industry, including the proteasome inhibitor salinosporamide A, which has entered the first phase of clinical trials as a cancer treatment (4). While the functions of some of the <i>Salinispora</i> metabolites are still unknown, these bacteria may offer researchers a source of new antibiotics and anticancer therapeutics.<br> | <br>Rather than designing synthetic drugs, some researchers are exploring natural sources for cancer treatments and as models for future synthetic treatments. One promising source includes the recently discovered <i>Salinispora</i> bacteria, belonging to the order <i>Actinomycetales</i> (also known as actinomycetes). This order includes many species that have contributed to today’s naturally occurring antibiotics. It was originally believed that the gram positive <i>Actinomycetales</i> only existed in soil habitats, and that any actinomycetes found in marine environments came from spores produced by terrestrial species (1). However, in search of new actinomycetes as possible dug sources, Jensen and Fenical of the Scripps Institution of Oceanography in the late 1980s discovered actinomycetes (originally called MAR1) that required seawater for growth (2). In 2002, the separate strains of marine origin were found in tropical and subtropical sediment, and classified under the genus <i>Salinispora</i>, belonging to the family <i>Micromonosporaceae</i>. Two species have been formally identified as <i>Salinispora arenicola</i> and <i>Salinispora tropica</i>, while a third species of <i>Salinispora pacifica</i> is being studied (2,3). The <i>Salinispora</i> bacteria have, like the rest of their order, a variety of secondary metabolites that may be useful in the pharmeceutical industry, including the proteasome inhibitor salinosporamide A, which has entered the first phase of clinical trials as a cancer treatment (4). While the functions of some of the <i>Salinispora</i> metabolites are still unknown, these bacteria may offer researchers a source of new antibiotics and anticancer therapeutics.<br> | ||
==<i>Salinispora</i>== | ==<i>Salinispora</i>== | ||
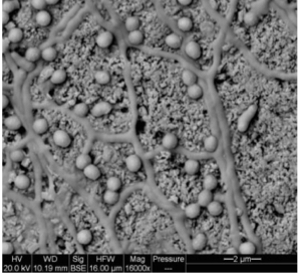
[[Image: | [[Image:Actinomycetes.png|thumb|300px|right| Figure 1. An actinomycete on a sand grain (SEM) with spores and filaments.(6) ]] | ||
<br> <i>Salinispora </i> are aerobic, Gram-positive, non-acid fast bacteria (5). This species is found in marine sediment both near the shoreline, as well as at a depth of 2000 meters. It is has also been cultured in association with sea grass and algae (1, 2, 6). It is believed that like the related terrestrial actinomycetes, <i>Salinispora </i> are involved in decomposing organic materials along the ocean floor (6). <i> Salinispora </i>form branched, non-fragmenting hyphae that are 0.25 to .5 um in diameter, with N-glycolated muramic acid and meso-diaminopimelic acid in their cell wall peptidoglycan (5). These hyphae have been seen growing away from the pieces of shells or sand grains to which the bacteria appear attached (6). Major sugars used within these bacteria are galactose, arabinose, and xylose. <i> Salinispora </i> do not have mycolic acids, but contain anteiso-, saturated, and iso-fatty acids (5). Salt water or a salt-supplemented growth medium is required for growth, a key distinction between <i> Salinispora </i> and its close relatives. <i>Salinispora </i> growth occurs at a pH of 7-12 and 10-30°C. | <br> <i>Salinispora </i> are aerobic, Gram-positive, non-acid fast bacteria (5). This species is found in marine sediment both near the shoreline, as well as at a depth of 2000 meters. It is has also been cultured in association with sea grass and algae (1, 2, 6). It is believed that like the related terrestrial actinomycetes, <i>Salinispora </i> are involved in decomposing organic materials along the ocean floor (6). <i> Salinispora </i>form branched, non-fragmenting hyphae that are 0.25 to .5 um in diameter, with N-glycolated muramic acid and meso-diaminopimelic acid in their cell wall peptidoglycan (5). These hyphae have been seen growing away from the pieces of shells or sand grains to which the bacteria appear attached (Figure 1) (6). Major sugars used within these bacteria are galactose, arabinose, and xylose. <i> Salinispora </i> do not have mycolic acids, but contain anteiso-, saturated, and iso-fatty acids (5). Salt water or a salt-supplemented growth medium is required for growth, a key distinction between <i> Salinispora </i> and its close relatives. <i>Salinispora </i> growth occurs at a pH of 7-12 and 10-30°C. Spores, 0.8 to 3.8 um in diameter, appear as darkened areas of cultured colonies, and are formed on short, spore bearing stalks known as sporophores. These spores are smooth, non-motile, and can be produced in clusters or individually (5). When cultured on nutrient rich media, the bacteria form orange and/or black colored colonies without the presence of aerial hyphae. On nutrient poor media, <i>Salinispora</i> form mycelia and produce diffusible orange, pink, dark brown, or black pigments (Figure 2) (5). | ||
[[Image:06-07SalinisporaTropica.jpg|thumb|300px|left| [http://ucsdnews.ucsd.edu/newsrel/science/06-07DiseaseFightingAgentsMA-.asp Figure 2. <i>Salinispora tropica</i> sequenced at the University of California-San Diego's Scripps Institution of Oceanography and Skaggs School of Pharmacy and Pharmaceutical Sciences. ]]]<i>Salinispora</i> is currently divided into three species as determined by 16S rRNA gene sequence analysis. No overlap in secondary metabolic products between any of the defined species has been recorded (7). <i>S. arenicola </i> was found from coarse sand in marine sediments in Guam, the Red Sea, the Bahamas, Palau, the Sea of Cortez, and the U.S. Virgin Islands (8). Though several strains of this species have been acquired in various marine locations, the species-specific secondary metabolites have remained constant, suggesting they may be important for survival and can be used as a phenotypic indicator (7). The <i>S. arenicola</i> genome has been sequenced, and contains 5,786,361 bp (9). Its optimum growth occurs at 20-28°C with 25-50% seawater. Energy for this species is obtained through L-proline, L-threonine, L-tyrosine, arbutin, and (+)-D-salicin (5). <i> S. tropica </i>has only been cultured from marine sediment collection sites in the Bahamas (8). It grows optimally at 15-28°C, and obtains energy from inulin and galactose. <i>S. pacifica </i>has been cultured from Palau, the Red Sea, and Guam, and is still being categorized. Though these bacteria are mostly obtained from marine sediment, <i>S. arenicola </i>and <i>S. pacifica </i>have been located in a sponge from the Great Barrier Reef, and S. pacifica has been identified with an ascidian in Fiji (7). | |||
This genus of marine bacteria produces an array of secondary metabolites. | This genus of marine bacteria produces an array of secondary metabolites. Researchers have identified and chemically characterized these compounds isolated from cellular extracts through UV spectra, molecular weights, and retention times from liquid chromatography-mass spectrometry. Identified compounds include, anticancer, antibiotic, antiviral, and anti-inflammatory agents. However, not all of the metabolites produced by the <i>Salinispora </i>species have a known biological activity, and of those that have demonstrated a useful biological effect not all have known molecular targets (8). <br> | ||
==Anticancer agents== | ==Anticancer agents== | ||
<br>Cancer is a major issue both in the United States and globally. In 2008, 565,650 Americans were predicted to die of cancer. However, between 1996 and 2003 the cancer survival rate raised to 66% as a result of more accurate diagnosis and effective treatments (9). One research institution in 2008, the National Cancer Institute, spent 4.83 billion dollars on cancer research (10). Discovering novel anticancer agents both synthetically and naturally have become increasingly important as many cancers do not respond to current treatments. | <br>Cancer is a major issue both in the United States and globally. In 2008, 565,650 Americans were predicted to die of cancer. However, between 1996 and 2003 the cancer survival rate raised to 66% as a result of more accurate diagnosis and effective treatments (9). One research institution in 2008, the National Cancer Institute, spent 4.83 billion dollars on cancer research (10). Discovering novel anticancer agents both synthetically and naturally have become increasingly important as many cancers do not respond to current treatments. Four potential anticancer agents have been isolated from the <i>Salinispora</i> genus: salinosporamide A, staurosporine, and saliniketal A and B. | ||
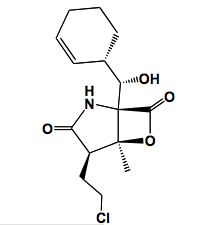
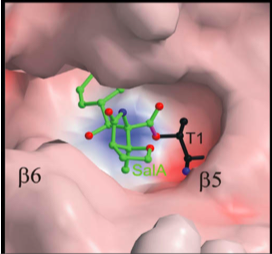
[[Image:Salinosporamide_A.png |thumb|300px|right| Figure 3. Salinosporamide A produced by <i>S. tropica</i>, a potential anticancer agent that inhibits proteasomes (9).]]Salinosporamide A is a secondary metabolite isolated from the culture broth of <i>S. tropica</i> that is currently being studied for use in cancer treatment. This compound can be isolated by adding a resin to a <i>Salinispora</i> culture, which is later removed by filtration, and eluted with acetone. Column chromatography and recrystallization methods are then used to collect salinosporamide A from the crude extract (11). Salinosporamide A is composed of connected γ-lactam and β-lactone rings as determined through NMR analysis (Figure 3) (12). It is very similar in structure to omuralide, which is an inhibitor of the 20S subunit of proteasomes, the multicatalytic protein complex responsible for degrading damaged or unusable proteins through peptide bond cleavage. From this structural similarity, researchers were able to determine that salinosproamide A also selectively inhibits the 20S proteasome. Within the 20S proteasome, there are three pairs of active proteolytic subunits (2). Salinosporamide A binds to, and inhibits all three activation sites through the formation of a cyclic ether with the catalytic site of the proteasome (Figure 4). This particular bond is believed to contribute to the irreversible binding of salinosporamide A to the 20S proteasome (2). Proteasomes have an important role in cancer development in that they regulate NFκB, a transcription factor that becomes constitutively activated in many cancers. NFκB controls genes that regulate proinflammatory cytokines, anti-apoptotic compounds, and cell-adhesion molecules (11).[[Image:20S_proteasome.png|thumb|300px|left| Figure 4. A model of salinosporamide A in the β5 subunit of a 20S proteasome (21 as cited in 9).]] | |||
Inhibiting proteasomes is currently a cancer treatment method through the antitumor drug bortezomib (Velcade). Salinosporamide A is potent, with an IC50 value of 1.3 nM compared to another candidate drug, omuralide, with 49 nM (13). The IC50 value indicates half of the drug concentration needed for maximum inhibition. The smaller the IC50 value, the less amount of the drug needed to produce maximum biological inhibition. Additionally, salinosporamide A is effective against multiple myeloma cells, cancerous plasma cells, that are Velcade-resistant (12). In order to manufacture salinosporamide A, passivation techniques needed to be used to prevent corrosion of industrial stainless steel fermentors by the saline solution needed for <i>S. tropica</i> metabolism (2). This is one of the first uses of saline fermentation to acquire a clinical trial drug. Though synthetically produced analogs have been examined, only the biologically produced salinosporamide A is currently being evaluated in clinical trials (2). | |||
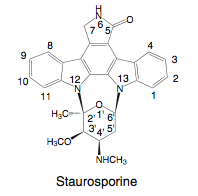
[[Image:Staurosporine.png|thumb|300px|right| Figure 5. Staurosporine produced by <i>S. arenicola</i>, a potential anticancer agent that inhibits protein serine/threonine and tyrosine kinases (14).]] | |||
<i>S. arenicola</i> also produces potential therapeutics, such as the well known staurosporine that is synthesized from two typtophan molecules. This compound was originally discovered in 1977 from the soil actinomycete <i>Streptomyces</i> (Figure 5) (14). It has been found in other actinomycete strains, cyanobacteria, and slime molds. Staurosporine inhibits protein serine/threonine and tyrosine kinases, which includes the protein kinase C (PKC). The phosphorylation activity of PKC is involved in many signaling pathways that regulate cell polarity, differentiation, cell cycle control, survival, and proliferation (15). PKC is also a receptor for tumor-enhancing phorbol esters. In cancer, PKC isozyme activity and expression levels are altered, becoming involved in apoptosis resistance, metastasis, enhanced proliferation, and angiogenesis (15). Thus, PKC isozymes have become targets for therapeutic cancer research. However, PKC does not specifically inhibit PKC, and researchers have further investigated or synthesized related compounds, such as 7-hydroxysporine, which has entered clinical investigations. | |||
Though much research has focused on the anticancer properties of staurosporine, this bacterial compound has other potentially useful properties, especially synthetically modified. Staurosporine is being investigated as a possible treatment for parasitic protists ( | Though much research has focused on the anticancer properties of staurosporine, this bacterial compound has other potentially useful properties, especially synthetically modified. Staurosporine is being investigated as a possible treatment for parasitic protists (14). Such treatments would inhibit protein kinases specific to the parasite. In <i>Plasmodium falciparum</i>, the parasite responsible for malaria, staurosporine inhibits a kinase involved in nuclear division, indicating another potential use for staurosporine. Staurosporine has been demonstrated to inhibit a protein kinase present in all known mycobacteria genomes, and can reduce the growth of <i>Mycobacterium tuberculosis</i>. Derivatives of staursporine have demonstrated antiviral qualities with the Epstien-Barr virus, cytomegalovirus, and human immunodeficiency virus. Staurosporine also has an inhibitory effect on topoisomerase complexes, which are targeted in some antibacterial and antitumor drugs (14). | ||
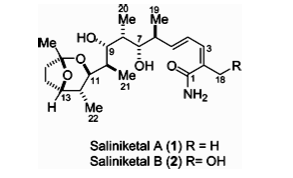
[[Image:SaliniketalAB.png|thumb|300px|left| Figure 6. Saliniketal A and Saliniketal B produced by <i>S. arenicola</i>, potential anticancer agents that inhibit ornithine dedecarboxylase (16).]] | |||
The secondary metabolites saliniketal A and B are bicyclic polyketides, containing alternating methylene and carbonyl groups, that have also been isolated from <i>S. arenicola</i> (Figure 6). Saliniketal B differs from Saliniketal A in that a allylic methyl is replaced by a methylene with an attached alcohol. These compounds have the potential to act as anti-cancer agents by inhibiting ornithine decarboxylase (ODC) (16). ODC is a limiting enzyme in polyamine biosynthesis, an important component in cell differentiation and growth (17). This enzyme decarboxylates ornithine to form the polyamine putrescine, and is found in eukaryotes and prokaryotes. ODC inhibitors are effective in both the eukaryotes and prokaryotes, though differences in amino acid sequences between these organisms exist. A bacteria producing an ODC inhibitor could hinder the growth of other bacteria and thus be evolutionarily advantageous. However, bacterial ODC inhibitors are being utilized in mammalian cells for a different purpose (16). Cancerous tissues have been found to contain more polyamines, allowing polyamine levels to be used in diagnosing cancer. Activated ODC has also been used to examine tumor progression. Blocking ODC can prevent tumor growth, which has made this enzyme a focus in preventative cancer research (17). | |||
==Antibiotics Anti-inflammatory Compounds== | |||
<br> | |||
Researchers found these compounds were ineffective when tested on T24 human bladder carcinoma cells. However, when concurrently used with TPA, a tumor promoter that promotes ODC activation, the saliniketals significantly inhibited ODC activity (16). Saliniketal A and B have IC50 values of 1.95 and 7.83 μg/mL when used in this manner. Chemical analysis has determined these compounds are of a different metabolic origin than rifamycin, an antibiotic compound produced by <i>S. arenicola</i> (16). <br> | |||
==Antibiotics & Anti-inflammatory Compounds== | |||
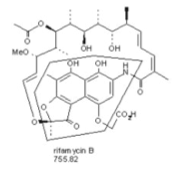
<br><i>S. arenicola</I> isolated from the sea sponge <i>Pseudoceratina clavata</i> produces rifamycin, an antibiotic that targets Gram-positive bacteria (Figure 7). The compound rifamycin was originally found in the soil actinomycetes <i>Amycolatopsis mediterranei</i>. Researchers discovered the production of rifamycin in this <i>Salinispora</i> species through finding a polyketide synthase gene cluster resembling that of <i>A. mediterranei</i>,which was amplified and analyzed through PCR. Liquid chromatography-tandem mass spectrometry was then used to detect the presence of rifamyin B and rifamycin SV in <i>Salinispora</i> cultures (18). The more active forms of rifamycin S and rifamycin SV originate from the compound rifamycin B, which can modified either naturally or synthetically. Rifamycin SV was the first rifamycin to be used clinically (18). Rifampicin, the semi-synthetic form of rifamycin, has been used to treat bacterial infections such as tuberculosis and chronic staphylococcal osteomyelitis, an inflammatory bone infection (18).[[Image:Rifamycin.png|thumb|500px|left| Figure 7. Rifamycin B produced by <i>S. arenicola</i>, an antibiotic affecting the DNA-dependent RNA polymerase (18).]] | |||
Rifamycin inhibits prokaryotic RNA synthesis through the DNA-dependent RNA polymerase (19). This inhibition occurs only during the initiation of RNA synthesis, with the complex becoming insensitive to rifamycin as a longer oligoribonucleotide chain is produced. Using rifampicin as a model, researchers discovered rifamycin binds to the β subunit in the DNA/RNA channel, physically preventing the elongation of the growing RNA chain. This causes the release of RNA chains containing only two or three nucleotides. However, the drug is suffering from developing bacterial resistance, and is usually administered in conjunction with another antibiotic. Resistance can occur through point mutations of the <i>rpoB</i> gene, which encodes the β subunit of the RNA polymerase, causing a decrease in the antibiotic’s ability to bind to the RNA polymerase. Different mutations lead to different degrees of rifamycin resistance. Other forms of resistance result from antibiotic modification, decreased sensitivity of the RNA polymerase to rifamycin, or inhibited uptake of the antibiotic (19). It is hoped that the discovery of a new rifamycin producing species will aid in developing new forms of rifamycin to combat the developing bacterial resistance (18).[[Image:CyclomarinA.png|thumb|300px|right| Figure 8. Cyclomarin A produced by <i>S. arenicola</i>, having anti-inflammatory and possible antiviral properties (20).]] | |||
<i>S. arenicola</i> also produces the anti-inflammatory metabolites cyclomarin A and C (Figure 8.). These agents were originally isolated from an aquatic streptomycete (20). Anti-inflamatory activity was measured through a phorbol ester-induced mouse ear edema assay, in which a mouse ear becomes inflamed after the application of phorbol ester. Swelling was reduced when cyclomarin A was administered topically or intraperitonially by 92% and 45% respectively (20). <i>S. arenicola</i> also produces cyclomarin D, which has a slight toxic effect on human colon carcinoma cells, as well as the cyclomarin derivatives of cydomarazine A and cydomarazine B. The cyclomarazines demonstrate inhibitory properties against vancomycin-resistant <i>Entrococcus faecium</i> at 13 ug/mL and methicillin-resistant <i>Staphylococcus areus</i> at 18 ug/mL. However, future studies must still be conducted to fully understand these compounds (20).<br> | |||
==Conclusion== | ==Conclusion== | ||
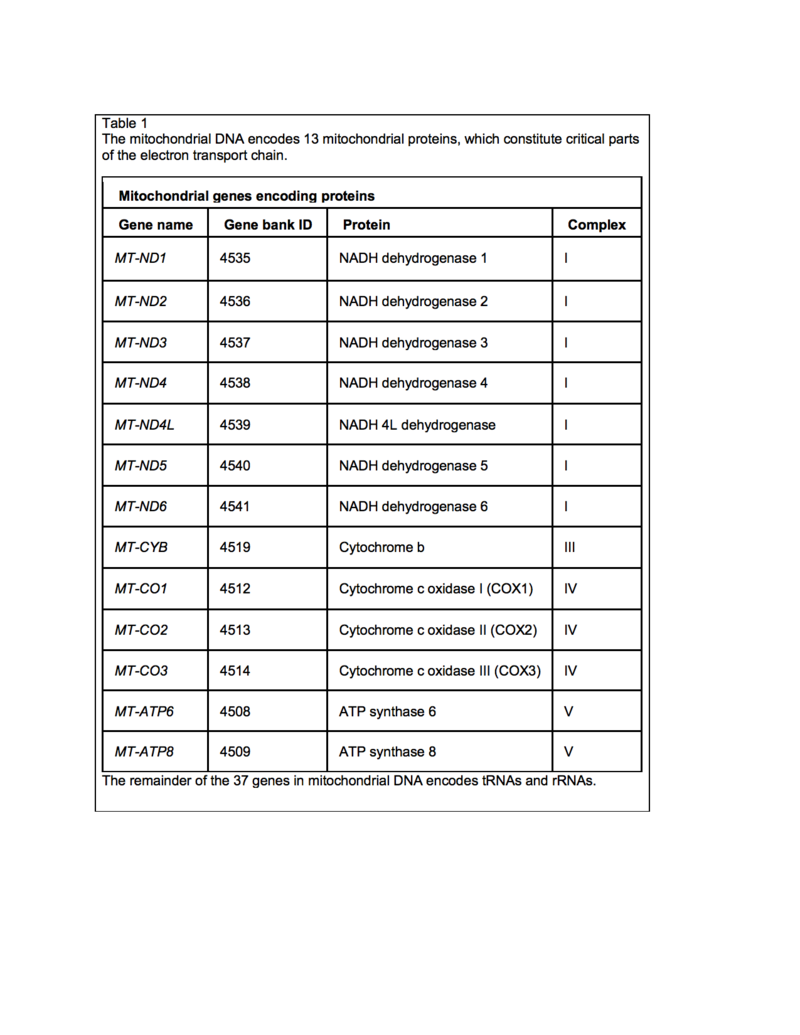
<br> | <br>Additional species and strains of <i>Salinispora</i> may yet be discovered. Much research is still needed before many of the previously described metabolites of <i>Salinispora</i> can enter clinical trials. Though some of these compounds have been found in other actinomycetes, a new source may provide additional derivatives or synthesizing pathways. The biological activity of many of the secondary metabolites isolated from the <I>Salinispora</i> species are still being investigated, including sporolide A from <i>S. tropica</i>, arenicolide A from <i>S. arenicola</i>, and cyanosporaside A and salinispyrone A from <i>S. pacifica</i> (Figure 9) (4). However, the discovery of this diverse set of metabolites highlights the potential resource marine bacteria may provide for potential novel therapeutics. <br> | ||
[[Image:Table.png|thumb|800px|center| Figure 9. Secondary metabolites of <i>Salinispora</i> and their biological activities (4).]] | |||
==References== | ==References== | ||
| Line 42: | Line 63: | ||
5. Maldonado, L.A., W. Fenical, P.R. Jensen, C.A. Kauffman, T.J. Mincer, A.C. Ward, A.T. Bull, and M. Goodfellow. 2005. Salinispora arenicola gen. nov., sp. nov. and Salinispora tropica sp. Nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. International Journal of Systematic and Evolutionary Microbiology. 55: 1759-1766. | 5. Maldonado, L.A., W. Fenical, P.R. Jensen, C.A. Kauffman, T.J. Mincer, A.C. Ward, A.T. Bull, and M. Goodfellow. 2005. Salinispora arenicola gen. nov., sp. nov. and Salinispora tropica sp. Nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. International Journal of Systematic and Evolutionary Microbiology. 55: 1759-1766. | ||
6. Jensen, P.R., P.G. Williams, D.C. Oh, L. Zeigler, and W. Fenical. 2007. Species-specific secondary metabolite production in marine Actinomycetes of the genus Salinispora. Applied and Experimental Microbiology. 73(4): 1146-1152. <br> | 6. Jensen, P.R., E. Gontang, C. Mafnas, T.J. Mincer, and W. Fenical. 2005. Culturable marine actinomycete diversity from tropical Pacific Ocean sediments. Environmental Microbiology. 7(7): 1039-1048. | ||
7. Jensen, P.R., and C. Mafnas. 2006. Biography of the marine actinomycete Salinispora. Environmental Microbiology. 8(11): 1881-1888. | |||
8. Jensen, P.R., P.G. Williams, D.C. Oh, L. Zeigler, and W. Fenical. 2007. Species-specific secondary metabolite production in marine Actinomycetes of the genus Salinispora. Applied and Experimental Microbiology. 73(4): 1146-1152. | |||
9. American Cancer Society. 2008. "Cancer Facts and Figures 2008". April 2009 http://www.cancer.org/downloads/STT/2008CAFFfinalsecured.pdf. | |||
10. National Cancer Institute. 13 March 2009. “Cancer Research Funding Fact Sheet.” U.S. National Institutes of Health. April 2009 http://www.cancer.gov/cancertopics/factsheet/NCI/research-funding. | |||
11. Feling, R.H., G.O. Buchanan. T.J. Mincer, C.A. Kauffman, P.R. Jensen, and W. Fenical. 2003. Salinosporamide A: A highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus <i>Salinospora</i>. Angew. Chem. Int. Ed. 42(3): 355-257. | |||
12. Williams, P.G., G.O. Buchanan, R.H. Feling, C.A. Kauffman, P.R. Jensen, and W. Fenical. 2005. New Cytotoxic Salinosporamides form the Marine Actinomycete Salinispora tropica. Journal of Organic Chemistry. 70: 6196-6203. | |||
13. Moore, B.S., A.S. Eustáquio, and R.P. McGlinchey. 2008. Advances in and applications of proteasome inhibitors. Current Opinion in Chemical Biology. 12: 434-440. | |||
14. Nakano, H., and S. Ômura. 2009. Chemical biology of natural indolocarbazole products: 30 years since the discovery of staurosporine. The Journal of Anitbiotics. 62: 17-26. | |||
15. Fields, A.P., and R.P. Regala. 2007. Protein kinase C: human oncogene, prognostic marker, and therapeutic target. Pharmacological Research. 55: 487-497. | |||
16. Williams, P.G., R.N. Asolkar, T. Kondratyuk, J.M. Pezzuto, P.R. Jensen, and W. Fenical. 2007. Saliniketals A and B, bicyclic polyketides from the Marine Actinomycete Salinispora arenicola. Journal of Natural Products. 70: 83-88. | |||
17. Deng, W., X. Jiang, Y. Mei, J. Sun, R. Ma, X. Liu, H. Sun, H. Tian, and X. Sun. 2008. Role of ornithine decarboxylase in breast cancer. Acta Biochimica et Biophysica Sinica. 40(3): 235-243. | |||
18. Kim, T.K., A.K. Hewavitharana, P.N. Shaw, and J.A. Fuerst. 2006. Discovery of a new source of rifamycin antibiotics in marine sponge actinobacteria by phylogenetic prediction. Applied and Environmental Microbiology. 72(3): 2118-2125. | |||
19. Floss, H.G., and T.W. Yu. 2005. Rifamycin-mode of action, resistance, and biosynthesis. Chemical Reviews. 105: 621-632. | |||
20. Renner, M.K., Y.C. Shen, X.C. Cheng, P.R. Jensen, W. Frankmoelle, C.A. Kauffman, W. Fenical, E. Lobkovsky, and J. Clardy. 1999. Cyclomarins A-C, new antiflammatory cyclic peptides produced by a marine bacterium (Streptomyces sp.). Journal of the American Chemical Society. 121: 11273-11276. | |||
21. Groll, M., R. Huber, and B.C.M. Potts. 2006. Crystal structures of salinosporamide A (NPR-0052) and B (NPI-0047) in complex with the 20S proteasome reveal important consequences of beta-lactone ring opening and a mechanism for irreversible binding. Journal of American Chemical Society. 128(15): 5136-5141.<br> | |||
Edited by a student of [mailto:slonczewski@kenyon.edu Joan Slonczewski] for [http://biology.kenyon.edu/courses/biol238/biol238syl09.html BIOL 238 Microbiology], 2009, [http://www.kenyon.edu/index.xml Kenyon College]. | Edited by a student of [mailto:slonczewski@kenyon.edu Joan Slonczewski] for [http://biology.kenyon.edu/courses/biol238/biol238syl09.html BIOL 238 Microbiology], 2009, [http://www.kenyon.edu/index.xml Kenyon College]. | ||
Latest revision as of 20:14, 10 August 2010
Christina Kucher
Introduction
Rather than designing synthetic drugs, some researchers are exploring natural sources for cancer treatments and as models for future synthetic treatments. One promising source includes the recently discovered Salinispora bacteria, belonging to the order Actinomycetales (also known as actinomycetes). This order includes many species that have contributed to today’s naturally occurring antibiotics. It was originally believed that the gram positive Actinomycetales only existed in soil habitats, and that any actinomycetes found in marine environments came from spores produced by terrestrial species (1). However, in search of new actinomycetes as possible dug sources, Jensen and Fenical of the Scripps Institution of Oceanography in the late 1980s discovered actinomycetes (originally called MAR1) that required seawater for growth (2). In 2002, the separate strains of marine origin were found in tropical and subtropical sediment, and classified under the genus Salinispora, belonging to the family Micromonosporaceae. Two species have been formally identified as Salinispora arenicola and Salinispora tropica, while a third species of Salinispora pacifica is being studied (2,3). The Salinispora bacteria have, like the rest of their order, a variety of secondary metabolites that may be useful in the pharmeceutical industry, including the proteasome inhibitor salinosporamide A, which has entered the first phase of clinical trials as a cancer treatment (4). While the functions of some of the Salinispora metabolites are still unknown, these bacteria may offer researchers a source of new antibiotics and anticancer therapeutics.
Salinispora
Salinispora are aerobic, Gram-positive, non-acid fast bacteria (5). This species is found in marine sediment both near the shoreline, as well as at a depth of 2000 meters. It is has also been cultured in association with sea grass and algae (1, 2, 6). It is believed that like the related terrestrial actinomycetes, Salinispora are involved in decomposing organic materials along the ocean floor (6). Salinispora form branched, non-fragmenting hyphae that are 0.25 to .5 um in diameter, with N-glycolated muramic acid and meso-diaminopimelic acid in their cell wall peptidoglycan (5). These hyphae have been seen growing away from the pieces of shells or sand grains to which the bacteria appear attached (Figure 1) (6). Major sugars used within these bacteria are galactose, arabinose, and xylose. Salinispora do not have mycolic acids, but contain anteiso-, saturated, and iso-fatty acids (5). Salt water or a salt-supplemented growth medium is required for growth, a key distinction between Salinispora and its close relatives. Salinispora growth occurs at a pH of 7-12 and 10-30°C. Spores, 0.8 to 3.8 um in diameter, appear as darkened areas of cultured colonies, and are formed on short, spore bearing stalks known as sporophores. These spores are smooth, non-motile, and can be produced in clusters or individually (5). When cultured on nutrient rich media, the bacteria form orange and/or black colored colonies without the presence of aerial hyphae. On nutrient poor media, Salinispora form mycelia and produce diffusible orange, pink, dark brown, or black pigments (Figure 2) (5).
Salinispora is currently divided into three species as determined by 16S rRNA gene sequence analysis. No overlap in secondary metabolic products between any of the defined species has been recorded (7). S. arenicola was found from coarse sand in marine sediments in Guam, the Red Sea, the Bahamas, Palau, the Sea of Cortez, and the U.S. Virgin Islands (8). Though several strains of this species have been acquired in various marine locations, the species-specific secondary metabolites have remained constant, suggesting they may be important for survival and can be used as a phenotypic indicator (7). The S. arenicola genome has been sequenced, and contains 5,786,361 bp (9). Its optimum growth occurs at 20-28°C with 25-50% seawater. Energy for this species is obtained through L-proline, L-threonine, L-tyrosine, arbutin, and (+)-D-salicin (5). S. tropica has only been cultured from marine sediment collection sites in the Bahamas (8). It grows optimally at 15-28°C, and obtains energy from inulin and galactose. S. pacifica has been cultured from Palau, the Red Sea, and Guam, and is still being categorized. Though these bacteria are mostly obtained from marine sediment, S. arenicola and S. pacifica have been located in a sponge from the Great Barrier Reef, and S. pacifica has been identified with an ascidian in Fiji (7).
This genus of marine bacteria produces an array of secondary metabolites. Researchers have identified and chemically characterized these compounds isolated from cellular extracts through UV spectra, molecular weights, and retention times from liquid chromatography-mass spectrometry. Identified compounds include, anticancer, antibiotic, antiviral, and anti-inflammatory agents. However, not all of the metabolites produced by the Salinispora species have a known biological activity, and of those that have demonstrated a useful biological effect not all have known molecular targets (8).
Anticancer agents
Cancer is a major issue both in the United States and globally. In 2008, 565,650 Americans were predicted to die of cancer. However, between 1996 and 2003 the cancer survival rate raised to 66% as a result of more accurate diagnosis and effective treatments (9). One research institution in 2008, the National Cancer Institute, spent 4.83 billion dollars on cancer research (10). Discovering novel anticancer agents both synthetically and naturally have become increasingly important as many cancers do not respond to current treatments. Four potential anticancer agents have been isolated from the Salinispora genus: salinosporamide A, staurosporine, and saliniketal A and B.
Salinosporamide A is a secondary metabolite isolated from the culture broth of S. tropica that is currently being studied for use in cancer treatment. This compound can be isolated by adding a resin to a Salinispora culture, which is later removed by filtration, and eluted with acetone. Column chromatography and recrystallization methods are then used to collect salinosporamide A from the crude extract (11). Salinosporamide A is composed of connected γ-lactam and β-lactone rings as determined through NMR analysis (Figure 3) (12). It is very similar in structure to omuralide, which is an inhibitor of the 20S subunit of proteasomes, the multicatalytic protein complex responsible for degrading damaged or unusable proteins through peptide bond cleavage. From this structural similarity, researchers were able to determine that salinosproamide A also selectively inhibits the 20S proteasome. Within the 20S proteasome, there are three pairs of active proteolytic subunits (2). Salinosporamide A binds to, and inhibits all three activation sites through the formation of a cyclic ether with the catalytic site of the proteasome (Figure 4). This particular bond is believed to contribute to the irreversible binding of salinosporamide A to the 20S proteasome (2). Proteasomes have an important role in cancer development in that they regulate NFκB, a transcription factor that becomes constitutively activated in many cancers. NFκB controls genes that regulate proinflammatory cytokines, anti-apoptotic compounds, and cell-adhesion molecules (11).
Inhibiting proteasomes is currently a cancer treatment method through the antitumor drug bortezomib (Velcade). Salinosporamide A is potent, with an IC50 value of 1.3 nM compared to another candidate drug, omuralide, with 49 nM (13). The IC50 value indicates half of the drug concentration needed for maximum inhibition. The smaller the IC50 value, the less amount of the drug needed to produce maximum biological inhibition. Additionally, salinosporamide A is effective against multiple myeloma cells, cancerous plasma cells, that are Velcade-resistant (12). In order to manufacture salinosporamide A, passivation techniques needed to be used to prevent corrosion of industrial stainless steel fermentors by the saline solution needed for S. tropica metabolism (2). This is one of the first uses of saline fermentation to acquire a clinical trial drug. Though synthetically produced analogs have been examined, only the biologically produced salinosporamide A is currently being evaluated in clinical trials (2).
S. arenicola also produces potential therapeutics, such as the well known staurosporine that is synthesized from two typtophan molecules. This compound was originally discovered in 1977 from the soil actinomycete Streptomyces (Figure 5) (14). It has been found in other actinomycete strains, cyanobacteria, and slime molds. Staurosporine inhibits protein serine/threonine and tyrosine kinases, which includes the protein kinase C (PKC). The phosphorylation activity of PKC is involved in many signaling pathways that regulate cell polarity, differentiation, cell cycle control, survival, and proliferation (15). PKC is also a receptor for tumor-enhancing phorbol esters. In cancer, PKC isozyme activity and expression levels are altered, becoming involved in apoptosis resistance, metastasis, enhanced proliferation, and angiogenesis (15). Thus, PKC isozymes have become targets for therapeutic cancer research. However, PKC does not specifically inhibit PKC, and researchers have further investigated or synthesized related compounds, such as 7-hydroxysporine, which has entered clinical investigations.
Though much research has focused on the anticancer properties of staurosporine, this bacterial compound has other potentially useful properties, especially synthetically modified. Staurosporine is being investigated as a possible treatment for parasitic protists (14). Such treatments would inhibit protein kinases specific to the parasite. In Plasmodium falciparum, the parasite responsible for malaria, staurosporine inhibits a kinase involved in nuclear division, indicating another potential use for staurosporine. Staurosporine has been demonstrated to inhibit a protein kinase present in all known mycobacteria genomes, and can reduce the growth of Mycobacterium tuberculosis. Derivatives of staursporine have demonstrated antiviral qualities with the Epstien-Barr virus, cytomegalovirus, and human immunodeficiency virus. Staurosporine also has an inhibitory effect on topoisomerase complexes, which are targeted in some antibacterial and antitumor drugs (14).
The secondary metabolites saliniketal A and B are bicyclic polyketides, containing alternating methylene and carbonyl groups, that have also been isolated from S. arenicola (Figure 6). Saliniketal B differs from Saliniketal A in that a allylic methyl is replaced by a methylene with an attached alcohol. These compounds have the potential to act as anti-cancer agents by inhibiting ornithine decarboxylase (ODC) (16). ODC is a limiting enzyme in polyamine biosynthesis, an important component in cell differentiation and growth (17). This enzyme decarboxylates ornithine to form the polyamine putrescine, and is found in eukaryotes and prokaryotes. ODC inhibitors are effective in both the eukaryotes and prokaryotes, though differences in amino acid sequences between these organisms exist. A bacteria producing an ODC inhibitor could hinder the growth of other bacteria and thus be evolutionarily advantageous. However, bacterial ODC inhibitors are being utilized in mammalian cells for a different purpose (16). Cancerous tissues have been found to contain more polyamines, allowing polyamine levels to be used in diagnosing cancer. Activated ODC has also been used to examine tumor progression. Blocking ODC can prevent tumor growth, which has made this enzyme a focus in preventative cancer research (17).
Researchers found these compounds were ineffective when tested on T24 human bladder carcinoma cells. However, when concurrently used with TPA, a tumor promoter that promotes ODC activation, the saliniketals significantly inhibited ODC activity (16). Saliniketal A and B have IC50 values of 1.95 and 7.83 μg/mL when used in this manner. Chemical analysis has determined these compounds are of a different metabolic origin than rifamycin, an antibiotic compound produced by S. arenicola (16).
Antibiotics & Anti-inflammatory Compounds
S. arenicola isolated from the sea sponge Pseudoceratina clavata produces rifamycin, an antibiotic that targets Gram-positive bacteria (Figure 7). The compound rifamycin was originally found in the soil actinomycetes Amycolatopsis mediterranei. Researchers discovered the production of rifamycin in this Salinispora species through finding a polyketide synthase gene cluster resembling that of A. mediterranei,which was amplified and analyzed through PCR. Liquid chromatography-tandem mass spectrometry was then used to detect the presence of rifamyin B and rifamycin SV in Salinispora cultures (18). The more active forms of rifamycin S and rifamycin SV originate from the compound rifamycin B, which can modified either naturally or synthetically. Rifamycin SV was the first rifamycin to be used clinically (18). Rifampicin, the semi-synthetic form of rifamycin, has been used to treat bacterial infections such as tuberculosis and chronic staphylococcal osteomyelitis, an inflammatory bone infection (18).
Rifamycin inhibits prokaryotic RNA synthesis through the DNA-dependent RNA polymerase (19). This inhibition occurs only during the initiation of RNA synthesis, with the complex becoming insensitive to rifamycin as a longer oligoribonucleotide chain is produced. Using rifampicin as a model, researchers discovered rifamycin binds to the β subunit in the DNA/RNA channel, physically preventing the elongation of the growing RNA chain. This causes the release of RNA chains containing only two or three nucleotides. However, the drug is suffering from developing bacterial resistance, and is usually administered in conjunction with another antibiotic. Resistance can occur through point mutations of the rpoB gene, which encodes the β subunit of the RNA polymerase, causing a decrease in the antibiotic’s ability to bind to the RNA polymerase. Different mutations lead to different degrees of rifamycin resistance. Other forms of resistance result from antibiotic modification, decreased sensitivity of the RNA polymerase to rifamycin, or inhibited uptake of the antibiotic (19). It is hoped that the discovery of a new rifamycin producing species will aid in developing new forms of rifamycin to combat the developing bacterial resistance (18).
S. arenicola also produces the anti-inflammatory metabolites cyclomarin A and C (Figure 8.). These agents were originally isolated from an aquatic streptomycete (20). Anti-inflamatory activity was measured through a phorbol ester-induced mouse ear edema assay, in which a mouse ear becomes inflamed after the application of phorbol ester. Swelling was reduced when cyclomarin A was administered topically or intraperitonially by 92% and 45% respectively (20). S. arenicola also produces cyclomarin D, which has a slight toxic effect on human colon carcinoma cells, as well as the cyclomarin derivatives of cydomarazine A and cydomarazine B. The cyclomarazines demonstrate inhibitory properties against vancomycin-resistant Entrococcus faecium at 13 ug/mL and methicillin-resistant Staphylococcus areus at 18 ug/mL. However, future studies must still be conducted to fully understand these compounds (20).
Conclusion
Additional species and strains of Salinispora may yet be discovered. Much research is still needed before many of the previously described metabolites of Salinispora can enter clinical trials. Though some of these compounds have been found in other actinomycetes, a new source may provide additional derivatives or synthesizing pathways. The biological activity of many of the secondary metabolites isolated from the Salinispora species are still being investigated, including sporolide A from S. tropica, arenicolide A from S. arenicola, and cyanosporaside A and salinispyrone A from S. pacifica (Figure 9) (4). However, the discovery of this diverse set of metabolites highlights the potential resource marine bacteria may provide for potential novel therapeutics.
References
1. Mincer, T.J., P.R. Jensen, C.A. Kauffman, and W. Fenical. 2002. Widespread and Persistent Populations of a Major New Marine Actinomycete Taxon in Ocean Sediments. Applied and Experimental Microbiology. 68: 5005-5011.
2. Fenical, W., P.R. Jensen, M.A. Palladino, K.S. Lam, G.K. Lloyd, and B.C. Potts. 2009. Discovery and development of the anticancer agent salinosporamide A (NPI-0052). Bioorganic & Medicinal Chemistry. 17: 2175-2180.
3. Udwary, D.W., L. Zeigler, R.N. Asolkar, V. Singan, A. Lapidus, W. Fenical, P.R. Jensen, and B.S. Moore. 2007. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. PNAS. 104 (25): 10376-10381.
4. Jensen, P.R., P.G. Williams, D.C. Oh, L. Zeigler, and W. Fenical. Species-Specific Secondary Metabolite Production in Marine Actinomycetes of the Genus Salinispora. 2006. Applied and Experimental Microbiology. 73(4): 1146-1152.
5. Maldonado, L.A., W. Fenical, P.R. Jensen, C.A. Kauffman, T.J. Mincer, A.C. Ward, A.T. Bull, and M. Goodfellow. 2005. Salinispora arenicola gen. nov., sp. nov. and Salinispora tropica sp. Nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. International Journal of Systematic and Evolutionary Microbiology. 55: 1759-1766.
6. Jensen, P.R., E. Gontang, C. Mafnas, T.J. Mincer, and W. Fenical. 2005. Culturable marine actinomycete diversity from tropical Pacific Ocean sediments. Environmental Microbiology. 7(7): 1039-1048.
7. Jensen, P.R., and C. Mafnas. 2006. Biography of the marine actinomycete Salinispora. Environmental Microbiology. 8(11): 1881-1888.
8. Jensen, P.R., P.G. Williams, D.C. Oh, L. Zeigler, and W. Fenical. 2007. Species-specific secondary metabolite production in marine Actinomycetes of the genus Salinispora. Applied and Experimental Microbiology. 73(4): 1146-1152.
9. American Cancer Society. 2008. "Cancer Facts and Figures 2008". April 2009 http://www.cancer.org/downloads/STT/2008CAFFfinalsecured.pdf.
10. National Cancer Institute. 13 March 2009. “Cancer Research Funding Fact Sheet.” U.S. National Institutes of Health. April 2009 http://www.cancer.gov/cancertopics/factsheet/NCI/research-funding.
11. Feling, R.H., G.O. Buchanan. T.J. Mincer, C.A. Kauffman, P.R. Jensen, and W. Fenical. 2003. Salinosporamide A: A highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angew. Chem. Int. Ed. 42(3): 355-257.
12. Williams, P.G., G.O. Buchanan, R.H. Feling, C.A. Kauffman, P.R. Jensen, and W. Fenical. 2005. New Cytotoxic Salinosporamides form the Marine Actinomycete Salinispora tropica. Journal of Organic Chemistry. 70: 6196-6203.
13. Moore, B.S., A.S. Eustáquio, and R.P. McGlinchey. 2008. Advances in and applications of proteasome inhibitors. Current Opinion in Chemical Biology. 12: 434-440.
14. Nakano, H., and S. Ômura. 2009. Chemical biology of natural indolocarbazole products: 30 years since the discovery of staurosporine. The Journal of Anitbiotics. 62: 17-26.
15. Fields, A.P., and R.P. Regala. 2007. Protein kinase C: human oncogene, prognostic marker, and therapeutic target. Pharmacological Research. 55: 487-497.
16. Williams, P.G., R.N. Asolkar, T. Kondratyuk, J.M. Pezzuto, P.R. Jensen, and W. Fenical. 2007. Saliniketals A and B, bicyclic polyketides from the Marine Actinomycete Salinispora arenicola. Journal of Natural Products. 70: 83-88.
17. Deng, W., X. Jiang, Y. Mei, J. Sun, R. Ma, X. Liu, H. Sun, H. Tian, and X. Sun. 2008. Role of ornithine decarboxylase in breast cancer. Acta Biochimica et Biophysica Sinica. 40(3): 235-243.
18. Kim, T.K., A.K. Hewavitharana, P.N. Shaw, and J.A. Fuerst. 2006. Discovery of a new source of rifamycin antibiotics in marine sponge actinobacteria by phylogenetic prediction. Applied and Environmental Microbiology. 72(3): 2118-2125.
19. Floss, H.G., and T.W. Yu. 2005. Rifamycin-mode of action, resistance, and biosynthesis. Chemical Reviews. 105: 621-632.
20. Renner, M.K., Y.C. Shen, X.C. Cheng, P.R. Jensen, W. Frankmoelle, C.A. Kauffman, W. Fenical, E. Lobkovsky, and J. Clardy. 1999. Cyclomarins A-C, new antiflammatory cyclic peptides produced by a marine bacterium (Streptomyces sp.). Journal of the American Chemical Society. 121: 11273-11276.
21. Groll, M., R. Huber, and B.C.M. Potts. 2006. Crystal structures of salinosporamide A (NPR-0052) and B (NPI-0047) in complex with the 20S proteasome reveal important consequences of beta-lactone ring opening and a mechanism for irreversible binding. Journal of American Chemical Society. 128(15): 5136-5141.
Edited by a student of Joan Slonczewski for BIOL 238 Microbiology, 2009, Kenyon College.