Lipothrixviridae: Difference between revisions
No edit summary |
No edit summary |
||
| (17 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
{{Curated}} | |||
{{Viral Biorealm Family}} | |||
[[Image:lipothrixviridae.gif|thumb|right|Lipothrixviridae. (source: [http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/38000000.htm ICTVdb Descriptions])]] | |||
==Baltimore Classification== | ==Baltimore Classification== | ||
| Line 7: | Line 12: | ||
===Genera=== | ===Genera=== | ||
Alphalipothrixvirus, Betalipothrixvirus, Gammalipothrixvirus | ''Alphalipothrixvirus'', ''Betalipothrixvirus'', ''Gammalipothrixvirus'' | ||
==Description and Significance== | ==Description and Significance== | ||
Lipothrixviridae is a family crenarchaeal viruses. It is by far the most diverse family of crenarchaeal viruses, with six isolates divided into three genera: ''Alphalipothrixvirus'', ''Betalipothrixvirus'', and ''Gammalipothrixvirus''. ''Alphalipothrixvirus'' contains TTV1, TTV2, and TTV3, isolated from acidic hot springs Iceland. ''Betalipothrixvirus'' contains SIFV, also isolated in Iceland. Finally, ''Gammalipothrixvirus'' is represented by AFV1, isolated from Yellowstone National Park. (sources: [http://www.pubmedcentral.gov/articlerender.fcgi?tool=pubmed&pubmedid=15901711 Häring et al.], Ortmann et al.) | |||
==Genome Structure== | |||
The genome of Lipothrixviridae is not segmented and contains a single molecule of linear double-stranded DNA. The complete genome is 16000 nucleotides long. (source: [http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/38000000.htm ICTVdB]) | |||
==Virion Structure of a Lipothrixviridae== | |||
The virions of Lipothrixviridae consist of an envelope and a nucleocapsid. The virus capsid is enveloped and the virions are rod-shaped, rigid and have protrusions extending from the core through the envelope that arise asymmetrically from both ends. The virions measure 38 nm in diameter and are 410 nm long with a tight fitting membrane. The envelope has no surface projections. The capsid is elongated and exhibits helical symmetry and the core is helical. (source: [http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/38000000.htm ICTVdB]) | |||
== | ==Reproductive Cycle of a Lipothrixviridae in a Host Cell== | ||
The basic replication cycle of crenarchaeal viruses, including Lipothrixviruses, has yet to be determined. There are, however, some trends which have been reported. It is thought that members of the Lipothrixviridae family, as well as members of the [[Fuselloviridae]], [[Rudiviridae]], and [[Guttaviridae]] families, associate with host cells by tail fibers which are present on one or both ends of the virion. Some viruses integrate their genome into the host cell's chromosome, while others maintain their genomes as extrachromosomal elements. | |||
Assembly and release of Lipothrixviruses, as well as most known crenarchaeal viruses, do not require cell lysis. Most crenarchaeal viruses appear to set up chronic infections, either continually producing virus particles or doing so in short events resulting in growth inhibition. These long-term chronic infections are thought to be an adaptation to the extremely hot and acidic environments crenarchaeal viruses inhabit. (source: Ortmann et al.) | |||
== | ==Viral Ecology & Pathology== | ||
The Lipothrixviridae infect both acidophilic thermophiles (such as [[Sulfolobus]] and Acidianus species) as well as thermophiles from more neutral environments (such as ''Thermoproteus tenax''). Like most crenarchaeal viruses, the Lipothrixviridae set up chronic infections rather than lytic infections (i.e., the host cell is not lysed). (source: Ortmann et al.) | |||
==References== | |||
== | [http://www.pubmedcentral.gov/articlerender.fcgi?tool=pubmed&pubmedid=15901711 Häring et al. "Structure and Genome Organization of AFV2, a Novel Archaeal Lipothrixvirus with Unusual Terminal and Core Structures." ''Journal of Bacteriology'' 187.11 (2005): 3855–3858.] | ||
[http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/38000000.htm ICTVdB - The Universal Virus Database, version 3. http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/] | |||
Ortmann et al. "Hot crenarchaeal viruses reveal deep evolutionary connections." ''Nature Reviews Microbiology'' 4 (2006): 520-528. | |||
Latest revision as of 00:29, 8 August 2010
A Viral Biorealm page on the family Lipothrixviridae
Baltimore Classification
Higher order taxa
Viruses; dsDNA viruses, no RNA stage; Lipothrixviridae
Genera
Alphalipothrixvirus, Betalipothrixvirus, Gammalipothrixvirus
Description and Significance
Lipothrixviridae is a family crenarchaeal viruses. It is by far the most diverse family of crenarchaeal viruses, with six isolates divided into three genera: Alphalipothrixvirus, Betalipothrixvirus, and Gammalipothrixvirus. Alphalipothrixvirus contains TTV1, TTV2, and TTV3, isolated from acidic hot springs Iceland. Betalipothrixvirus contains SIFV, also isolated in Iceland. Finally, Gammalipothrixvirus is represented by AFV1, isolated from Yellowstone National Park. (sources: Häring et al., Ortmann et al.)
Genome Structure
The genome of Lipothrixviridae is not segmented and contains a single molecule of linear double-stranded DNA. The complete genome is 16000 nucleotides long. (source: ICTVdB)
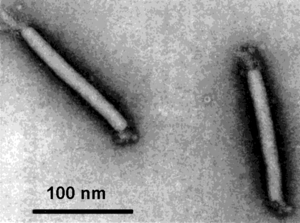
Virion Structure of a Lipothrixviridae
The virions of Lipothrixviridae consist of an envelope and a nucleocapsid. The virus capsid is enveloped and the virions are rod-shaped, rigid and have protrusions extending from the core through the envelope that arise asymmetrically from both ends. The virions measure 38 nm in diameter and are 410 nm long with a tight fitting membrane. The envelope has no surface projections. The capsid is elongated and exhibits helical symmetry and the core is helical. (source: ICTVdB)
Reproductive Cycle of a Lipothrixviridae in a Host Cell
The basic replication cycle of crenarchaeal viruses, including Lipothrixviruses, has yet to be determined. There are, however, some trends which have been reported. It is thought that members of the Lipothrixviridae family, as well as members of the Fuselloviridae, Rudiviridae, and Guttaviridae families, associate with host cells by tail fibers which are present on one or both ends of the virion. Some viruses integrate their genome into the host cell's chromosome, while others maintain their genomes as extrachromosomal elements.
Assembly and release of Lipothrixviruses, as well as most known crenarchaeal viruses, do not require cell lysis. Most crenarchaeal viruses appear to set up chronic infections, either continually producing virus particles or doing so in short events resulting in growth inhibition. These long-term chronic infections are thought to be an adaptation to the extremely hot and acidic environments crenarchaeal viruses inhabit. (source: Ortmann et al.)
Viral Ecology & Pathology
The Lipothrixviridae infect both acidophilic thermophiles (such as Sulfolobus and Acidianus species) as well as thermophiles from more neutral environments (such as Thermoproteus tenax). Like most crenarchaeal viruses, the Lipothrixviridae set up chronic infections rather than lytic infections (i.e., the host cell is not lysed). (source: Ortmann et al.)
References
ICTVdB - The Universal Virus Database, version 3. http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/
Ortmann et al. "Hot crenarchaeal viruses reveal deep evolutionary connections." Nature Reviews Microbiology 4 (2006): 520-528.
