Intestinal Microflora and Antibiotic Resistance: Difference between revisions
Slonczewski (talk | contribs) |
|||
| (56 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{{Curated}} | |||
==Basic Attributes of Gut Microflora== | ==Basic Attributes of Gut Microflora== | ||
The microflora of the human gastrointestinal (GI) tract is so extensive and integral to the proper functioning of the digestive system that it has been characterized as an additional organ of the human body (1). It is estimated that the microfloral community consists of 500 -1000 distinct species of bacteria in a single person; collectively, their population is 10 times the number of their host’s body cells (2). The intestine provides a suitable niche for many species of bacteria, as it remains at a stable temperature and is replete with bioavailable carbon, nitrogen, and solute sources of nutrition (7). The suite of microbes is acquired primarily during infancy, since the GI tract of a fetus is sterile during development. Bacteria are initially transferred to the infant during the delivery process, then continually from the immediate environment and from contact with its mother and other adults. Escherichia and Streptococcus are the first to colonize the GI tract, typically followed by | [[Image:4441009a-f1.2.jpg|thumb|330px|right|"The human gut and (inset) a scanning electron micrograph of part of the small intestine, with some bacterial inhabitants picked out in green." From [http://www.nature.com/nature/journal/v444/n7122/fig_tab/4441009a_F1.html Nature.com].]] | ||
The microflora of the human gastrointestinal (GI) tract is so extensive and integral to the proper functioning of the digestive system that it has been characterized as an additional organ of the human body (1). It is estimated that the microfloral community consists of 500 -1000 distinct species of bacteria in a single person; collectively, their population is 10 times the number of their host’s body cells (2). The intestine provides a suitable niche for many species of bacteria, as it remains at a stable temperature and is replete with bioavailable carbon, nitrogen, and solute sources of nutrition (7). The suite of microbes is acquired primarily during infancy, since the GI tract of a fetus is sterile during development. Bacteria are initially transferred to the infant during the delivery process, then continually from the immediate environment and from contact with its mother and other adults. Escherichia and Streptococcus are the first to colonize the GI tract, typically followed by <i>Bifidobacteria</i>, <i>Staphylococci</i>, <i>Lactobacilli</i>, <i>Micrococci</i>, and <i>Propionibacteria</i>. As the infant matures, it is continually exposed to bacteria, principally by the digestion of food, and the makeup of its intestinal microbiome changes dramatically (2, 5). The dynamic nature of the microfloral community entails shifting of community species composition over time and variation in composition among individuals. However, the predominant taxa of bacteria in the gut are fairly consistent. <i>Firmicutes</i> and <i>Bacteroides</i> are the most prevalent phyla by far; populations of <i>Proteobacteria</i>, <i>Actinobacteria</i>, <i>Fusobacteria</i>, and <i>Verrucobacteria</i> are frequently established as well (2, 6). | |||
====Functions of Intestinal Microflora==== | ====Functions of Intestinal Microflora==== | ||
The functions performed by gut bacterial include regulation of intestine epithelial development, maintenance of | The functions performed by gut bacterial include regulation of intestine epithelial development, maintenance of mucosal homeostasis and repair, improved absorption of nutrients from food, and contributions to the innate immune system (1, 2, 3). <br> | ||
Colonization of Bacteriodes induces the upregulation of sodium/glucose transporters; improved utilization of glucose from metabolism allows the intestine to grow and repair itself faster and more efficiently (3). | <br><B>Development</B><br> | ||
Colonization of <i>Bacteriodes</i> induces the upregulation of sodium/glucose transporters; improved utilization of glucose from metabolism allows the intestine to grow and repair itself faster and more efficiently (3). The colonization of various other species of bacteria also contribute to the epithelium by causing the mucosal layer to significantly increase in depth. This growth contributes to both the successful growth and maintenence of flora colonies and the defenses against potentially pathogenic bacteria (16). Research has indicated that the period of weaning in mice (which represents a period of microfloral diversification) is accompanied by the expansion of a villous capillary network that is absent in germ-free mice; also, growth of this network can be induced with the addition of <i>B. thetaiotaomicron</i>. This suggests more effective transport of nutrients in the intestines of mammals with healthy microflora (3). | |||
<br><B>Digestion</B><br> | |||
These functions are all highly relevant to medicine, as a deeper understanding of how bacteria influence these systems will yield more sophisticated solutions to conditions affecting the body. | The metabolism of some complex polysaccharides, lipids, and other compounds is only possible in the presence of bacterial species. Many bacteria specialize in the metabolism of certain compounds, such as <i>B. thetaiotaomicron</i>: well over half of the genome of this species consists of machinery that breaks down highly-structured polysaccharides. These bacteria have also been shown to differentiate gene expression to improve the efficiency of their glycan foraging behavior, which likely allows the host access to nutrients from a greater variety of food types. Germ-free mice require 20-30% more calories to maintain their metabolic rate than mice with normal gut flora; they also display digestive issues of varying severity when introduced to more complex food. In general, lipids are ineffectively metabolized and stored in germ-free mice, leading to fluctuating weight. When these mice were colonized with a full suite of floral bacteria, they gained an average of 60% more body fat and exhibited a 14% increase in leptin, a known appetite suppressant and regulator of glucose and insulin levels (3, 14). | ||
An important factor to the effective functioning of the human microbiome is the movement of genetic material. Genes are exchanged both | |||
<br><B>Innate Immunity</B><br> | |||
The innate immune system is reinforced by the microbiome by myriad means, most of which are likely undocumented. Known responses to intestinal conditions include the stimulation of antibacterial proteins following an invasion event, triggering of inflammation after moderate injury, and maintaining protection from pathogens by interacting with toll-like receptor cells. These are discussed in section 3. | |||
<br><br> These functions are all highly relevant to medicine, as a deeper understanding of how bacteria influence these systems will yield more sophisticated solutions to conditions affecting the body. | |||
An important factor to the effective functioning of the human microbiome is the movement of genetic material. Genes are exchanged both between bacteria species, via lateral gene transfer, and among bacteria and their host. The presence of 233 documented human proteins that have homologs only in bacteria hints at the extent to which human development and function has historically been strongly influenced by bacteria (2). The transfer of genetic material among bacterial species affects human hosts in more immediate and clinically relevant ways, including, notably, influencing response to antibiotics (4). | |||
==Studying Human Microflora== | ==Studying Human Microflora== | ||
Characterizing the diversity and function of the microbiome is notoriously difficult. Most gut bacteria cannot be cultured due to their very particular nutritional requirements and sensitivity to oxygen. While many species in the proximal region of the gut are facultatively aerobic, tolerance of oxygen decreases when approaching the distal region of the intesine (2). Because keeping bacterial communities intact in vitro is so challenging, previous studies may be significantly biased in their analyses of composition and function. The use of phenotypic characteristics to classify bacteria (which was utilized until only recently) also leads to an incomplete and oversimplified resolution of species. More modern techniques to evaluate bacterial communities and their evolutionary lineages involve whole-genome sequencing and a strong emphasis on rRNA (1, 4, 6). The ubiquity of 16S strands in prokaryotic lineages makes them a natural choice for elucidating diversity and evolutionary histories of gut microbiota. rRNA 16S, which is located on the 30S subunit of bacterial ribosomes, contains regions that are highly conserved, variable, and highly variable; the conserved “signature” sequences indicate evolutionary history of a particular taxonomic group with precision. A number of techniques have been employed to sequence 16S rRNA, including sequencing of cloned amplicons, direct sequencing, TRFLP, dot-blot hybridization, shotgun sequencing, and quantitative PCR (or real-time PCR) (9). Cloned amplicon sequencing is common, due to its moderately high resolution and ability to clone the full length of the 16S strand. The 16S sequences are treated with primers that bind to these conserved regions and amplified with standard PCR techniques, so that the resulting clones may be identified and catalogued (8). Though all of these sequencing techniques are powerful, they are inherently limited by the method of sampling. Fecal samples are commonly used to assess bacterial composition, but do not accurately represent the flora community. Mucosal tissue samples directly from the intestine are preferable, but are more invasive and expensive to obtain (1). Studies utilizing these molecular techniques consistently identify great numbers of novel species (with benchmark similarity values of 95% or greater), indicating efforts to characterize the gut flora have been insufficient. It is estimated in review literature that only 19% of the adult flora has been identified, and only 8% of the | [[File:rRNA.jpg|350px|thumb|left|Organization of the 16S rRNA sequence, used in phylogenetic analyses of bacterial communities. From [http://www.pnas.org/content/102/46/16620/F1.expansion.html PNAS]]] | ||
Characterizing the diversity and function of the microbiome is notoriously difficult. Most gut bacteria cannot be cultured due to their very particular nutritional requirements and sensitivity to oxygen. While many species in the proximal region of the gut are facultatively aerobic, tolerance of oxygen decreases when approaching the distal region of the intesine (2). Because keeping bacterial communities intact in vitro is so challenging, previous studies may be significantly biased in their analyses of composition and function. The use of phenotypic characteristics to classify bacteria (which was utilized until only recently) also leads to an incomplete and oversimplified resolution of species. More modern techniques to evaluate bacterial communities and their evolutionary lineages involve whole-genome sequencing and a strong emphasis on rRNA (1, 4, 6). | |||
===16S rRNA and Bacterial Characterization=== | |||
The ubiquity of 16S strands in prokaryotic lineages makes them a natural choice for elucidating diversity and evolutionary histories of gut microbiota. rRNA 16S, which is located on the 30S subunit of bacterial ribosomes, contains regions that are highly conserved, variable, and highly variable; the conserved “signature” sequences indicate evolutionary history of a particular taxonomic group with precision. A number of techniques have been employed to sequence 16S rRNA, including sequencing of cloned amplicons, direct sequencing, TRFLP, dot-blot hybridization, shotgun sequencing, and quantitative PCR (or real-time PCR) (9). Cloned amplicon sequencing is common, due to its moderately high resolution and ability to clone the full length of the 16S strand. The 16S sequences are treated with primers that bind to these conserved regions and amplified with standard PCR techniques, so that the resulting clones may be identified and catalogued (8). | |||
===Types of Sampling=== | |||
Though all of these sequencing techniques are powerful, they are inherently limited by the method of sampling. Fecal samples are commonly used to assess bacterial composition, but do not accurately represent the flora community. Mucosal tissue samples directly from the intestine are preferable, but are more invasive and expensive to obtain (1). Studies utilizing these molecular techniques consistently identify great numbers of novel species (with benchmark similarity values of 95% or greater), indicating efforts to characterize the gut flora have been insufficient. It is estimated in review literature that only 19% of the adult flora has been identified, and only 8% of the elderly flora (9). Due to the density of the microbial system and the complex interactions among species and between bacteria and the host, many researchers consider the gut to be its own ecosystem. This perspective has influenced the analysis of the data output from these sequencing projects: many ecological measures of richness, abundance, and growth are used to predict bacterial dynamics (1, 17). | |||
<br> | |||
<br> | |||
<br> | |||
==The Gut Microbiome and the Defenses of the Intestine== | ==The Gut Microbiome and the Defenses of the Intestine== | ||
Since the intestine is such an attractive habitat for microbes, it must employ many types of defenses to protect the body from invasion of pathogen microbes. Many of these defense mechanisms are intricately related to a normal population of gut microflora. Microflora inhabit the mucosal layer of the intestine, which is a gel-like substance secreted by goblet cells onto the lumen-facing surface. Compared in function to a biofilm, this mucus layer can either trap or prevent the adhesion of potentially pathogenic organisms (depending on the pathogen's physiology and adaptive ability). The invasion of exogenous bacteria, both Gram-positive and Gram-negative, stimulates the secretion of mucus, strengthening this form of defense (16). Indigenous bacteria must be specially adapted to dwell in this layer. The intestine constantly “checks on” the population of microflora using toll-like receptors. Two pathways have been identified as ways TLRs protect microflora: either TLRs constitutively express protective factors in response to concentration of microbial products, or protection factors are produced in response to physical or chemical damage to the epithelium (15). Evidence that intestinal microbiota have a distinct and crucial effect on the stimulation of mucus production is presented in a study that compared goblet cells of germ-free mice to conventional mice. The size of the goblet cells were significantly augmented in conventional specimens, but more importantly the mucosal layer was up to two times as thick. The structural components of gut flora bacteria that trigger this thickening effect remain unknown (17). | [[Image:Largeintestine.jpg|thumb|400px|right|Histological image of the human large intestine, with the mucosal and submucosal layers clearly visible. From [http://www.columbia.edu/cu/biology/courses/w2501/histology.html Columbia University Biology Department].]]Since the intestine is such an attractive habitat for microbes, it must employ many types of defenses to protect the body from invasion of pathogen microbes. Many of these defense mechanisms are intricately related to a normal population of gut microflora. Microflora inhabit the mucosal layer of the intestine, which is a gel-like substance secreted by goblet cells onto the lumen-facing surface. Compared in function to a biofilm, this mucus layer can either trap or prevent the adhesion of potentially pathogenic organisms (depending on the pathogen's physiology and adaptive ability). The invasion of exogenous bacteria, both Gram-positive and Gram-negative, stimulates the secretion of mucus, strengthening this form of defense (16). Indigenous bacteria must be specially adapted to dwell in this layer, and use a variety of structural means to “bury” themselves in the protective mucus and reproduce successfully. The intestine constantly “checks on” the population of microflora using toll-like receptors. Two pathways have been identified as ways TLRs protect microflora: either TLRs constitutively express protective factors in response to concentration of microbial products, or protection factors are produced in response to physical or chemical damage to the epithelium (15). Evidence that intestinal microbiota have a distinct and crucial effect on the stimulation of mucus production is presented in a study that compared goblet cells of germ-free mice to conventional mice. The size of the goblet cells were significantly augmented in conventional specimens, but more importantly the mucosal layer was up to two times as thick. The structural components of gut flora bacteria that trigger this thickening effect remain unknown (17). | ||
Resistance to antibiotics is a necessary attribute of bacterial populations dwelling in the human gut. The epithelium of the intestine expresses a variety of antimicrobial agents as a first-line defense of the immune system. These peptides are either expressed constitutively or induced by inflammatory mediators (10). Thus, in order to flourish in the human gut commensal bacteria must be recognized by toll-like receptors and other mediators as well as be resistant to these comparatively non-specific antimicrobial agents. Pathogenic bacteria are recognized by their structural attributes by means of complex pathways, then are destroyed by inflammation or the antimicrobial peptides (11). The ability to distinguish between commensal bacteria and potentially pathogenic bacteria is crucial to the continued function of the microflora “organ” as well as the immune system as a whole. | Resistance to antibiotics is a necessary attribute of bacterial populations dwelling in the human gut. The epithelium of the intestine expresses a variety of antimicrobial agents as a first-line defense of the immune system. These peptides are either expressed constitutively or induced by inflammatory mediators and other messenger components (10). In fact, expression of antimicrobial proteins can be induced by the flora themselves: the protein angiogenin-4, which kills Gram-positive bacteria, is produced in response to signals from specific indigenous bacteria (3). Thus, in order to flourish in the human gut commensal bacteria must be recognized by toll-like receptors and other mediators as well as be resistant to these comparatively non-specific antimicrobial agents. Pathogenic bacteria are recognized by their structural attributes by means of complex pathways, then are destroyed by inflammation or the antimicrobial peptides (11). The ability to distinguish between commensal bacteria and potentially pathogenic bacteria is crucial to the continued function of the microflora “organ” as well as the immune system as a whole. | ||
==Effects of the Use of Antibiotics on Microbe Populations== | ==Effects of the Use of Antibiotics on Microbe Populations== | ||
The advent of broad-spectrum antibiotics has upset the delicate balance between commensal bacteria and the antimicrobial substances produced by the intestine. Antibiotics can be ingested either deliberately (in prescribed medications) or accidently (in meat from animals treated extensively with antibiotics in industrial farm settings). The prescription of antibiotics as the “first line” of defense for a wide range of symptoms that may not actually be bacterial in origin may lead to serious | The advent of broad-spectrum antibiotics has upset the delicate balance between commensal bacteria and the antimicrobial substances produced by the intestine. Antibiotics can be ingested either deliberately (in prescribed medications) or accidently (in meat from animals treated extensively with antibiotics in industrial farm settings). The prescription of antibiotics as the “first line” of defense for a wide range of symptoms that may not actually be bacterial in origin may lead to serious impacts on gut flora. Since broad-spectrum antibiotics are generally manufactured to be non-specific, vast numbers of bacteria in the gut are destroyed following administration of the drug (12). | ||
[[Image:Levaquin250mg.jpg|thumb|250px|left|A 250 mg dosage of levofloaxin (brand name Levaquin), a commonly prescribed broad-spectrum antibiotic. From [http://myhealth.ucsd.edu/library/healthguide/en-us/drugguide/topic.asp?hwid=d04109a1 The UCSD Healthwise Guide].]] | |||
===Loss of Microfloral Benefits=== | |||
The immediate effects of losing microflora function include, predictably, a decreased ability to metabolize certain carbohydrate and lipid structures that make up a significant portion of the diet (13), regulation of fat storage (14), and loss of antimicrobial structures expressed by many microflora species (2). These effects vary in duration from weeks to several months, depending on the health of the host and the extent to which flora was destroyed. Individuals vary in their response to antibiotic usage due to the variability of their bacterial colonies (further obscuring studies attempting to determine makeup of the gut microbiome); also, some taxa seem to be more vulnerable to antibiotic damage than others. Gut bacteria taxa that seem to suffer disproportionally include <i>Firmicutes</i>, <i>Bifidobacteria</i>, <i>Collinsella</i>, and <i>Ruminococcus</i>; <i>Proteobacteria</i> appears to be more resistant and shows transient dominance after treatment, despite making up only 1% of a healthy person's flora (4, 17). These results suggest that the spread of newly dominant species to utilize available nutrients may reduce the ability of normally distributed flora to regenerate, as well as fundamentally changing the biochemistry of the GI tract. It has also been demonstrated that Salmonella can opportunistically colonize fecal samples following treatment with ciprofloxacin, up to a threshold concentration of antibiotic – this suggests new vulnerabilities to pathogen attack as well (19). Multiple treatment episodes for recurrent infections or, ironically, antibiotic-resistant pathogens may also influence the ability of gut flora to return to their pre-drug status. | |||
===Recolonizing Bacteria After Treatment=== | |||
Methods for replenishing flora faster include the oral administration of probiotic supplements containing known concentrations of commensal bacteria (7) and conserving a sample of a person's own fecal matter, extracting the bacteria, and redepositing it either orally or through the colon. This could be preferable to using generic cultured bacteria, since the introduction of new bacteria could trigger an immune response. The methods of delivery each have advantages: oral delivery is less invasive, while colon delivery is faster and reduces bacteria loss. While still in development, this method is a promising means to mitigate antibiotic damage to gut flora (Personal Correspondence, [http://gi.duke.edu/modules/dukefaculty/viewDetails.php?d=liddl001&t=1 Dr. Rodger Liddle, Duke University]). | |||
==The Gut as a Reservoir for Antibiotic Resistance== | ==The Gut as a Reservoir for Antibiotic Resistance== | ||
A regimen of antibiotic drugs introduces an artificial selection gradient in the human body for | A regimen of antibiotic drugs introduces an artificial selection gradient in the human body for antibiotic resistance in microflora populations. Since antibiotic resistance genes tend to be encoded on transposable elements such as plasmids, they are readily transferred among populations via lateral gene transfer. The bacteria that receive the plasmids conferring resistance are at a distinct evolutionary advantage, regardless of other benefits they provide the host. Several studies have concluded that ermB and tetQ, which provide resistance to erythromycin and tetracycline, have increased substantially both in occurrence in the gut and the number of species in which they are present. This emphasis on resistance as a fitness factor represents a shift from historical means of keeping bacterial populations in balance. The “cocktail” application of several antibiotics at once to treat conditions such as stomach ulcers (caused by pathogenic bacteria <i>Helicobacter pylori</i>) may exacerbate the selection gradient as well. <br> | ||
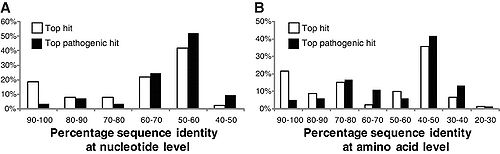
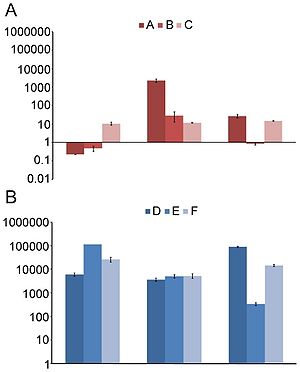
[[File:325_1128_F1.jpg|500px|thumb|left|"Distributions of (A) nucleotide identities and (B) amino acid identities for 93 resistance genes identified from DNA extracted directly from saliva and fecal samples to the most similar resistance gene from any organism (white bars) as well as the most similar resistance gene harbored by a pathogenic isolate (black bars) in GenBank. Two resistance genes with no significant similarity to sequences in GenBank are not shown." From [http://www.sciencemag.org/cgi/content/full/325/5944/1128 Sommer, et al.]]][[File:ermB.jpg|300px|thumb|right| "erm(B) abundance over time. | |||
The normalized fold increase of erm(B) compared to day 0 in community DNA extracted from fecal samples for controls (A–C) not receiving any treatment (A) and patients (D–F) receiving antibiotics (B). Each bar graph represents the mean and standard error of the normalized expression of erm(B) compared to 16S. Normalization was carried out as previously been described [8]." From [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2844414/ Jakobsson, et al.]]] Sommer, et al. (2009) studied fecal samples to determine the abundance and distribution of antibiotic resistance factors in microflora. They extracted DNA directly from bacteria from the guts of people who had not been treated with antibiotics for a year, then inserted these sequences into E. coli via expression vectors. The bacteria were cultured and divided onto antibiotic-laden plates, such that each group was subjected to one of thirteen antibiotics. The researchers sequenced those bacteria that contained resistance factors to all thirteen drugs. In all, 95 new resistance inserts were identified. Phylogenetic analysis revealed the most likely origin of antibiotic resistance factors to be Bacteroides and Firmicutes; this is unsurprising considering their dominance in the gut. Of principle interest to this study was how closely related the inserts were to previous entries in GenBank, since the genetic distance between sequences might indicate rate of transfer (assuming a moderately regular rate of mutation). The average similarity factors extracted from human flora and the closest known gene was 69.5% on the nucleotide level, and 65.3% on the amino acid level. Only 22% had >90% similarity to benchmark factors. These results are highly pertinent to the general system, but are not resolved to the question of exchange among indigenous and exogenous species. When they narrowed their perspective and compared resistance sequences of intestinal flora and pathogenic bacteria, they found that nearly half of the factors were identical in sequence to human pathogen genes. Though a direct means of transmission of genetic material from pathogens to human floral cells has not yet been demonstrated, these results have important implications for the human immune system and the effectiveness of antibiotics (4). If pathogens that invade the body transfer plasmids containing resistance factors to floral cells, even if they are destroyed by the immune systems the factor will remain. This may be beneficial to conserving and regenerating flora after being treated with an antibiotic regimen, but transfer occurs in both directions. If genes conferring resistance are transferred to pathogenic species, different cocktails of antibiotics at higher concentrations will be prescribed to fight off the infection, damaging the flora in novel ways. <br> | |||
The data of Sommer, et al. are supported and expanded upon by the recent work of Jakobsson, et al., who studied antibiotic resistance in human microflora in the context of time. Time is an essential component to the study of microflora because the regeneration (or failure thereof) of flora will determine how a patient will respond to a pathogen or antibiotic in the future. Rate of flora regrowth varies by individual, so the potential to refine medical techniques to be patient-specific is dependent on time studies. This team utilized TRFLP methods to “fingerprint” floral DNA and determine short- and long-term changes in species composition over time. They found the community composition shifts radically after exposure to clarithromycin and metronidazole (another common combination for H. pylori), favoring some taxa over others. Intriguingly, their results indicated that at after four full years since treatment, the original bacterial composition of the gut was not replenished. Of equal importance is the persistence of ermB, the resistance factor to erythromycin. The factor had clearly remained and spread among multiple populations; the authors propose that <i>Enterococci</i> survived the antibiotics and distributed the factor by both asexual reproduction and lateral transfer. | |||
==Conclusions== | ==Conclusions== | ||
| Line 50: | Line 70: | ||
:15) [http://journals.ohiolink.edu/ejc/xml_ft.cgi/Magalhaes_Joao_G.html?issn=10445323&issue=v19i0002&article=106_tiebhtbtmfap#sec3 Magalhaes, J. et al. 2007. The intestinal epithelial barrier: How to distinguish between the microbial flora and pathogens. Seminars in Immunology 19(2): 106-115.] | :15) [http://journals.ohiolink.edu/ejc/xml_ft.cgi/Magalhaes_Joao_G.html?issn=10445323&issue=v19i0002&article=106_tiebhtbtmfap#sec3 Magalhaes, J. et al. 2007. The intestinal epithelial barrier: How to distinguish between the microbial flora and pathogens. Seminars in Immunology 19(2): 106-115.] | ||
:16) [http://www.ajcn.org/cgi/content/full/73/6/1131S Deplancke, B. et al. 2001. Microbial modulation of innate defense: goblet cells and the intestinal mucus layer. American Journal of Clinical Nutrition, 73(6): 1131S-1141S.] | :16) [http://www.ajcn.org/cgi/content/full/73/6/1131S Deplancke, B. et al. 2001. Microbial modulation of innate defense: goblet cells and the intestinal mucus layer. American Journal of Clinical Nutrition, 73(6): 1131S-1141S.] | ||
:17 [http://www.ncbi.nlm.nih.gov/pubmed/8726140 Kandori, H. et al. 1996. Histochemical, lectin-histochemical and morphometrical characteristics of intestinal goblet cells of germfree and conventional mice. Exp Anim. 45(2):155-60.] | :17) [http://www.ncbi.nlm.nih.gov/pubmed/8726140 Kandori, H. et al. 1996. Histochemical, lectin-histochemical and morphometrical characteristics of intestinal goblet cells of germfree and conventional mice. Exp Anim. 45(2):155-60.] | ||
:18 [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WPT-4DSW68X-3&_user=7774802&_coverDate=12%2F01%2F2004&_rdoc=1&_fmt=high&_orig=search&_sort=d&_docanchor=&view=c&_acct=C000062877&_version=1&_urlVersion=0&_userid=7774802&md5=1e56c09c4083b59c6831022a4f2bc98e Carman, R. et al. 2004. Ciprofloxacin at low levels disrupts colonization resistance of human fecal microflora growing in chemostats. Regulatory Toxicology and Pharmacology 40(3): 319-326.] | :18) [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WPT-4DSW68X-3&_user=7774802&_coverDate=12%2F01%2F2004&_rdoc=1&_fmt=high&_orig=search&_sort=d&_docanchor=&view=c&_acct=C000062877&_version=1&_urlVersion=0&_userid=7774802&md5=1e56c09c4083b59c6831022a4f2bc98e Carman, R. et al. 2004. Ciprofloxacin at low levels disrupts colonization resistance of human fecal microflora growing in chemostats. Regulatory Toxicology and Pharmacology 40(3): 319-326.] | ||
Edited by Allison Price, a student of [mailto:slonczewski@kenyon.edu Joan Slonczewski] for [http://biology.kenyon.edu/courses/biol238/biol238syl09.html BIOL 238 Microbiology], 2010, [http://www.kenyon.edu/index.xml Kenyon College]. | |||
<!-- Do not edit or remove this line-->[[Category:Pages edited by students of Joan Slonczewski at Kenyon College]] | |||
Latest revision as of 01:14, 31 July 2013
Basic Attributes of Gut Microflora
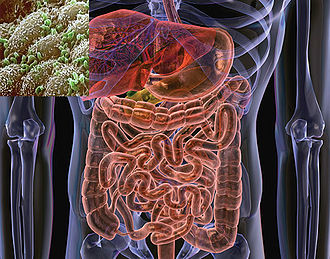
The microflora of the human gastrointestinal (GI) tract is so extensive and integral to the proper functioning of the digestive system that it has been characterized as an additional organ of the human body (1). It is estimated that the microfloral community consists of 500 -1000 distinct species of bacteria in a single person; collectively, their population is 10 times the number of their host’s body cells (2). The intestine provides a suitable niche for many species of bacteria, as it remains at a stable temperature and is replete with bioavailable carbon, nitrogen, and solute sources of nutrition (7). The suite of microbes is acquired primarily during infancy, since the GI tract of a fetus is sterile during development. Bacteria are initially transferred to the infant during the delivery process, then continually from the immediate environment and from contact with its mother and other adults. Escherichia and Streptococcus are the first to colonize the GI tract, typically followed by Bifidobacteria, Staphylococci, Lactobacilli, Micrococci, and Propionibacteria. As the infant matures, it is continually exposed to bacteria, principally by the digestion of food, and the makeup of its intestinal microbiome changes dramatically (2, 5). The dynamic nature of the microfloral community entails shifting of community species composition over time and variation in composition among individuals. However, the predominant taxa of bacteria in the gut are fairly consistent. Firmicutes and Bacteroides are the most prevalent phyla by far; populations of Proteobacteria, Actinobacteria, Fusobacteria, and Verrucobacteria are frequently established as well (2, 6).
Functions of Intestinal Microflora
The functions performed by gut bacterial include regulation of intestine epithelial development, maintenance of mucosal homeostasis and repair, improved absorption of nutrients from food, and contributions to the innate immune system (1, 2, 3).
Development
Colonization of Bacteriodes induces the upregulation of sodium/glucose transporters; improved utilization of glucose from metabolism allows the intestine to grow and repair itself faster and more efficiently (3). The colonization of various other species of bacteria also contribute to the epithelium by causing the mucosal layer to significantly increase in depth. This growth contributes to both the successful growth and maintenence of flora colonies and the defenses against potentially pathogenic bacteria (16). Research has indicated that the period of weaning in mice (which represents a period of microfloral diversification) is accompanied by the expansion of a villous capillary network that is absent in germ-free mice; also, growth of this network can be induced with the addition of B. thetaiotaomicron. This suggests more effective transport of nutrients in the intestines of mammals with healthy microflora (3).
Digestion
The metabolism of some complex polysaccharides, lipids, and other compounds is only possible in the presence of bacterial species. Many bacteria specialize in the metabolism of certain compounds, such as B. thetaiotaomicron: well over half of the genome of this species consists of machinery that breaks down highly-structured polysaccharides. These bacteria have also been shown to differentiate gene expression to improve the efficiency of their glycan foraging behavior, which likely allows the host access to nutrients from a greater variety of food types. Germ-free mice require 20-30% more calories to maintain their metabolic rate than mice with normal gut flora; they also display digestive issues of varying severity when introduced to more complex food. In general, lipids are ineffectively metabolized and stored in germ-free mice, leading to fluctuating weight. When these mice were colonized with a full suite of floral bacteria, they gained an average of 60% more body fat and exhibited a 14% increase in leptin, a known appetite suppressant and regulator of glucose and insulin levels (3, 14).
Innate Immunity
The innate immune system is reinforced by the microbiome by myriad means, most of which are likely undocumented. Known responses to intestinal conditions include the stimulation of antibacterial proteins following an invasion event, triggering of inflammation after moderate injury, and maintaining protection from pathogens by interacting with toll-like receptor cells. These are discussed in section 3.
These functions are all highly relevant to medicine, as a deeper understanding of how bacteria influence these systems will yield more sophisticated solutions to conditions affecting the body.
An important factor to the effective functioning of the human microbiome is the movement of genetic material. Genes are exchanged both between bacteria species, via lateral gene transfer, and among bacteria and their host. The presence of 233 documented human proteins that have homologs only in bacteria hints at the extent to which human development and function has historically been strongly influenced by bacteria (2). The transfer of genetic material among bacterial species affects human hosts in more immediate and clinically relevant ways, including, notably, influencing response to antibiotics (4).
Studying Human Microflora
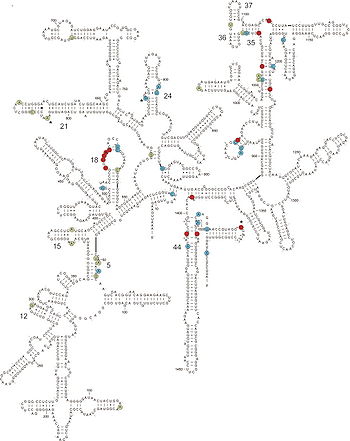
Characterizing the diversity and function of the microbiome is notoriously difficult. Most gut bacteria cannot be cultured due to their very particular nutritional requirements and sensitivity to oxygen. While many species in the proximal region of the gut are facultatively aerobic, tolerance of oxygen decreases when approaching the distal region of the intesine (2). Because keeping bacterial communities intact in vitro is so challenging, previous studies may be significantly biased in their analyses of composition and function. The use of phenotypic characteristics to classify bacteria (which was utilized until only recently) also leads to an incomplete and oversimplified resolution of species. More modern techniques to evaluate bacterial communities and their evolutionary lineages involve whole-genome sequencing and a strong emphasis on rRNA (1, 4, 6).
16S rRNA and Bacterial Characterization
The ubiquity of 16S strands in prokaryotic lineages makes them a natural choice for elucidating diversity and evolutionary histories of gut microbiota. rRNA 16S, which is located on the 30S subunit of bacterial ribosomes, contains regions that are highly conserved, variable, and highly variable; the conserved “signature” sequences indicate evolutionary history of a particular taxonomic group with precision. A number of techniques have been employed to sequence 16S rRNA, including sequencing of cloned amplicons, direct sequencing, TRFLP, dot-blot hybridization, shotgun sequencing, and quantitative PCR (or real-time PCR) (9). Cloned amplicon sequencing is common, due to its moderately high resolution and ability to clone the full length of the 16S strand. The 16S sequences are treated with primers that bind to these conserved regions and amplified with standard PCR techniques, so that the resulting clones may be identified and catalogued (8).
Types of Sampling
Though all of these sequencing techniques are powerful, they are inherently limited by the method of sampling. Fecal samples are commonly used to assess bacterial composition, but do not accurately represent the flora community. Mucosal tissue samples directly from the intestine are preferable, but are more invasive and expensive to obtain (1). Studies utilizing these molecular techniques consistently identify great numbers of novel species (with benchmark similarity values of 95% or greater), indicating efforts to characterize the gut flora have been insufficient. It is estimated in review literature that only 19% of the adult flora has been identified, and only 8% of the elderly flora (9). Due to the density of the microbial system and the complex interactions among species and between bacteria and the host, many researchers consider the gut to be its own ecosystem. This perspective has influenced the analysis of the data output from these sequencing projects: many ecological measures of richness, abundance, and growth are used to predict bacterial dynamics (1, 17).
The Gut Microbiome and the Defenses of the Intestine
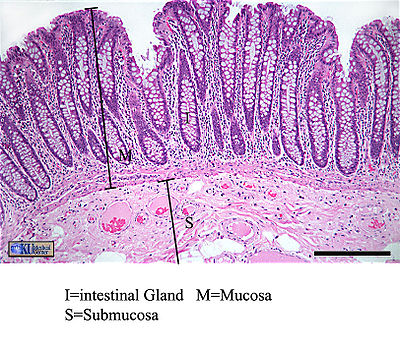
Since the intestine is such an attractive habitat for microbes, it must employ many types of defenses to protect the body from invasion of pathogen microbes. Many of these defense mechanisms are intricately related to a normal population of gut microflora. Microflora inhabit the mucosal layer of the intestine, which is a gel-like substance secreted by goblet cells onto the lumen-facing surface. Compared in function to a biofilm, this mucus layer can either trap or prevent the adhesion of potentially pathogenic organisms (depending on the pathogen's physiology and adaptive ability). The invasion of exogenous bacteria, both Gram-positive and Gram-negative, stimulates the secretion of mucus, strengthening this form of defense (16). Indigenous bacteria must be specially adapted to dwell in this layer, and use a variety of structural means to “bury” themselves in the protective mucus and reproduce successfully. The intestine constantly “checks on” the population of microflora using toll-like receptors. Two pathways have been identified as ways TLRs protect microflora: either TLRs constitutively express protective factors in response to concentration of microbial products, or protection factors are produced in response to physical or chemical damage to the epithelium (15). Evidence that intestinal microbiota have a distinct and crucial effect on the stimulation of mucus production is presented in a study that compared goblet cells of germ-free mice to conventional mice. The size of the goblet cells were significantly augmented in conventional specimens, but more importantly the mucosal layer was up to two times as thick. The structural components of gut flora bacteria that trigger this thickening effect remain unknown (17).
Resistance to antibiotics is a necessary attribute of bacterial populations dwelling in the human gut. The epithelium of the intestine expresses a variety of antimicrobial agents as a first-line defense of the immune system. These peptides are either expressed constitutively or induced by inflammatory mediators and other messenger components (10). In fact, expression of antimicrobial proteins can be induced by the flora themselves: the protein angiogenin-4, which kills Gram-positive bacteria, is produced in response to signals from specific indigenous bacteria (3). Thus, in order to flourish in the human gut commensal bacteria must be recognized by toll-like receptors and other mediators as well as be resistant to these comparatively non-specific antimicrobial agents. Pathogenic bacteria are recognized by their structural attributes by means of complex pathways, then are destroyed by inflammation or the antimicrobial peptides (11). The ability to distinguish between commensal bacteria and potentially pathogenic bacteria is crucial to the continued function of the microflora “organ” as well as the immune system as a whole.
Effects of the Use of Antibiotics on Microbe Populations
The advent of broad-spectrum antibiotics has upset the delicate balance between commensal bacteria and the antimicrobial substances produced by the intestine. Antibiotics can be ingested either deliberately (in prescribed medications) or accidently (in meat from animals treated extensively with antibiotics in industrial farm settings). The prescription of antibiotics as the “first line” of defense for a wide range of symptoms that may not actually be bacterial in origin may lead to serious impacts on gut flora. Since broad-spectrum antibiotics are generally manufactured to be non-specific, vast numbers of bacteria in the gut are destroyed following administration of the drug (12).
Loss of Microfloral Benefits
The immediate effects of losing microflora function include, predictably, a decreased ability to metabolize certain carbohydrate and lipid structures that make up a significant portion of the diet (13), regulation of fat storage (14), and loss of antimicrobial structures expressed by many microflora species (2). These effects vary in duration from weeks to several months, depending on the health of the host and the extent to which flora was destroyed. Individuals vary in their response to antibiotic usage due to the variability of their bacterial colonies (further obscuring studies attempting to determine makeup of the gut microbiome); also, some taxa seem to be more vulnerable to antibiotic damage than others. Gut bacteria taxa that seem to suffer disproportionally include Firmicutes, Bifidobacteria, Collinsella, and Ruminococcus; Proteobacteria appears to be more resistant and shows transient dominance after treatment, despite making up only 1% of a healthy person's flora (4, 17). These results suggest that the spread of newly dominant species to utilize available nutrients may reduce the ability of normally distributed flora to regenerate, as well as fundamentally changing the biochemistry of the GI tract. It has also been demonstrated that Salmonella can opportunistically colonize fecal samples following treatment with ciprofloxacin, up to a threshold concentration of antibiotic – this suggests new vulnerabilities to pathogen attack as well (19). Multiple treatment episodes for recurrent infections or, ironically, antibiotic-resistant pathogens may also influence the ability of gut flora to return to their pre-drug status.
Recolonizing Bacteria After Treatment
Methods for replenishing flora faster include the oral administration of probiotic supplements containing known concentrations of commensal bacteria (7) and conserving a sample of a person's own fecal matter, extracting the bacteria, and redepositing it either orally or through the colon. This could be preferable to using generic cultured bacteria, since the introduction of new bacteria could trigger an immune response. The methods of delivery each have advantages: oral delivery is less invasive, while colon delivery is faster and reduces bacteria loss. While still in development, this method is a promising means to mitigate antibiotic damage to gut flora (Personal Correspondence, Dr. Rodger Liddle, Duke University).
The Gut as a Reservoir for Antibiotic Resistance
A regimen of antibiotic drugs introduces an artificial selection gradient in the human body for antibiotic resistance in microflora populations. Since antibiotic resistance genes tend to be encoded on transposable elements such as plasmids, they are readily transferred among populations via lateral gene transfer. The bacteria that receive the plasmids conferring resistance are at a distinct evolutionary advantage, regardless of other benefits they provide the host. Several studies have concluded that ermB and tetQ, which provide resistance to erythromycin and tetracycline, have increased substantially both in occurrence in the gut and the number of species in which they are present. This emphasis on resistance as a fitness factor represents a shift from historical means of keeping bacterial populations in balance. The “cocktail” application of several antibiotics at once to treat conditions such as stomach ulcers (caused by pathogenic bacteria Helicobacter pylori) may exacerbate the selection gradient as well.
Sommer, et al. (2009) studied fecal samples to determine the abundance and distribution of antibiotic resistance factors in microflora. They extracted DNA directly from bacteria from the guts of people who had not been treated with antibiotics for a year, then inserted these sequences into E. coli via expression vectors. The bacteria were cultured and divided onto antibiotic-laden plates, such that each group was subjected to one of thirteen antibiotics. The researchers sequenced those bacteria that contained resistance factors to all thirteen drugs. In all, 95 new resistance inserts were identified. Phylogenetic analysis revealed the most likely origin of antibiotic resistance factors to be Bacteroides and Firmicutes; this is unsurprising considering their dominance in the gut. Of principle interest to this study was how closely related the inserts were to previous entries in GenBank, since the genetic distance between sequences might indicate rate of transfer (assuming a moderately regular rate of mutation). The average similarity factors extracted from human flora and the closest known gene was 69.5% on the nucleotide level, and 65.3% on the amino acid level. Only 22% had >90% similarity to benchmark factors. These results are highly pertinent to the general system, but are not resolved to the question of exchange among indigenous and exogenous species. When they narrowed their perspective and compared resistance sequences of intestinal flora and pathogenic bacteria, they found that nearly half of the factors were identical in sequence to human pathogen genes. Though a direct means of transmission of genetic material from pathogens to human floral cells has not yet been demonstrated, these results have important implications for the human immune system and the effectiveness of antibiotics (4). If pathogens that invade the body transfer plasmids containing resistance factors to floral cells, even if they are destroyed by the immune systems the factor will remain. This may be beneficial to conserving and regenerating flora after being treated with an antibiotic regimen, but transfer occurs in both directions. If genes conferring resistance are transferred to pathogenic species, different cocktails of antibiotics at higher concentrations will be prescribed to fight off the infection, damaging the flora in novel ways.
The data of Sommer, et al. are supported and expanded upon by the recent work of Jakobsson, et al., who studied antibiotic resistance in human microflora in the context of time. Time is an essential component to the study of microflora because the regeneration (or failure thereof) of flora will determine how a patient will respond to a pathogen or antibiotic in the future. Rate of flora regrowth varies by individual, so the potential to refine medical techniques to be patient-specific is dependent on time studies. This team utilized TRFLP methods to “fingerprint” floral DNA and determine short- and long-term changes in species composition over time. They found the community composition shifts radically after exposure to clarithromycin and metronidazole (another common combination for H. pylori), favoring some taxa over others. Intriguingly, their results indicated that at after four full years since treatment, the original bacterial composition of the gut was not replenished. Of equal importance is the persistence of ermB, the resistance factor to erythromycin. The factor had clearly remained and spread among multiple populations; the authors propose that Enterococci survived the antibiotics and distributed the factor by both asexual reproduction and lateral transfer.
Conclusions
The incredible diversity and richness of bacteria in the human gut is invaluable to our survival and, arguably, success as a species. Without the significant benefits to digestion and immune function conferred by bacteria, humans would be markedly less efficient and far more vulnerable to uncontrolled growth of pathogenic bacteria. Though the means to assess the specific types of bacteria and the benefits each population contributes is somewhat lacking, research has indubitably illustrated the significance of bacteria and the potentially harmful long-term effects of antibiotic drugs on microflora. The persistence of resistance in pathogenic bacteria is devastating in a hospital setting; the relatively recent phenomenon of gut bacteria acting as a source for resistance factors could have extremely harmful repercussions. Hopefully, future studies adding to the library of floral species and providing evidence for the destructive influence of over-prescribing antibiotics will convince practitioners to use antibiotics more judiciously.
Sources Cited
- 1) Eckburg, Paul B. et al. 2005. Diversity of the human intestinal microbial flora. Science 308: 1635-1638.
- 2) Sears, Cynthia L. 2005. A dynamic partnership: celebrating our gut flora. Science 11(5): 247-251.
- 3) Tsai, Franklin and Walter J. Coyle. 2009. The microbiome and obesity: Is obesity linked to our gut flora? Current Gastroenterology Reports 11(4): 307-314.
- 4) Sommer, O.A. Morten, et al. 2009. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325(5944): 1128-1131.
- 5) Mackie, Roderick I. et al. 1999. Development microbial ecology of the neonatal gastrointestinal tract. American Journal of Clinical Nutrition 69(5): 1035-1045.
- 6) Limdi, J.K., et al. 2005. Do probiotics have a therapeutic role in gastroenterology? World Journal of Gastroenterology 12(34): 5447-5457.
- 7) Engelbrektson, A. et al. 2009. Probiotics to minimize the disruption of faecal microbiota in healthy subjects undergoing antibiotic therapy. Journal of Medical Microbiology. 58(5) 663-670.
- 8) Blaut, M. et al. 2002. Molecular biological methods for studying the gut microbiota: the EU human gut flora project. British Journal of Nutrition 87(2): 203-211.
- 9) O'Toole, Paul W. and Marcus J Claesson. 2010. Gut microbiota: changes throughout the lifespan from infancy to elderly. International Dairy Journal 20: 281-291.
- 10) Harder, Jurgen et al. Isolation and Characterization of Human β-Defensin-3, a Novel Human Inducible Peptide Antibiotic. The Journal of Biological Chemistry 276: 5707-5713.
- 11) Rakoff-Nahoum, J. et al. 2001. Recognition of Commensal Microflora by Toll-Like Receptors Is Required for Intestinal Homeostasis. Cell 118(2): 229-241.
- 12) Jakobsson, H.E. et al. 2010. Short-Term Antibiotic Treatment Has Differing Long-Term Impacts on the Human Throat and Gut Microbiome. PLOS 5(3): e9836.
- 13)Wostmann, R. et al. 1983. Dietary intake, energy metabolism, and excretory losses of adult male germfree Wistar rats. Lab Animal Science 33(1): 46-50.
- 14) Backhed, F. et al. 2004. The gut microbiota as an environmental factor that regulates fat storage. PNAS 101(44): 15718-15723.
- 15) Magalhaes, J. et al. 2007. The intestinal epithelial barrier: How to distinguish between the microbial flora and pathogens. Seminars in Immunology 19(2): 106-115.
- 16) Deplancke, B. et al. 2001. Microbial modulation of innate defense: goblet cells and the intestinal mucus layer. American Journal of Clinical Nutrition, 73(6): 1131S-1141S.
- 17) Kandori, H. et al. 1996. Histochemical, lectin-histochemical and morphometrical characteristics of intestinal goblet cells of germfree and conventional mice. Exp Anim. 45(2):155-60.
- 18) Carman, R. et al. 2004. Ciprofloxacin at low levels disrupts colonization resistance of human fecal microflora growing in chemostats. Regulatory Toxicology and Pharmacology 40(3): 319-326.
Edited by Allison Price, a student of Joan Slonczewski for BIOL 238 Microbiology, 2010, Kenyon College.
