Cryptomonas: Difference between revisions
No edit summary |
|||
| (68 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | |||
[[Image:cryptomonas.jpg|frame|right| A Cryptomonas ovata phytoplankter from a sample collected at Lake Toolik (Alaska, USA). From [http://starcentral.mbl.edu/msr/rawdata/viewable/cryptomonas12_tkw.jpg micro*scope]]] | [[Image:cryptomonas.jpg|frame|right| A Cryptomonas ovata phytoplankter from a sample collected at Lake Toolik (Alaska, USA). From [http://starcentral.mbl.edu/msr/rawdata/viewable/cryptomonas12_tkw.jpg micro*scope]]] | ||
==Classification== | ==Classification== | ||
Eukaryota; Cryptophyta; Cryptomonadales; Cryptomonadaceae; Cryptomonas | Eukaryota; Cryptophyta; Cryptomonadales; Cryptomonadaceae; Cryptomonas | ||
| Line 7: | Line 7: | ||
{| | {| | ||
| height="10" bgcolor="#FFDF95" | | | height="10" bgcolor="#FFDF95" | | ||
''' | ''' ''Cryptomonas borealis'' [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=233190]''' | ||
|} | |||
{| | |||
| height="10" bgcolor="#FFDF95" | | |||
''' ''Cryptomonas commutata'' [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=400139]''' | |||
|} | |||
{| | |||
| height="10" bgcolor="#FFDF95" | | |||
''' ''Cryptomonas curvata'' [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=233186]''' | |||
|} | |||
{| | |||
| height="10" bgcolor="#FFDF95" | | |||
''' ''Cryptomonas erosa'' [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=400142]''' | |||
|} | |||
{| | |||
| height="10" bgcolor="#FFDF95" | | |||
''' ''Cryptomonas gyropyrenoidosa'' [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=233257]''' | |||
|} | |||
{| | |||
| height="10" bgcolor="#FFDF95" | | |||
''' ''Cryptomonas lundii'' [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=233258]''' | |||
|} | |||
{| | |||
| height="10" bgcolor="#FFDF95" | | |||
''' ''Cryptomonas loricata'' [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=400141]''' | |||
|} | |||
{| | |||
| height="10" bgcolor="#FFDF95" | | |||
''' ''Cryptomonas marssonii'' [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=233187]''' | |||
|} | |||
{| | |||
| height="10" bgcolor="#FFDF95" | | |||
''' ''Cryptomonas obovoidea'' [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=400143]''' | |||
|} | |} | ||
''Cryptomonas | {| | ||
| height="10" bgcolor="#FFDF95" | | |||
''' ''Cryptomonas ovata'' [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=70450]''' | |||
|} | |||
{| | |||
| height="10" bgcolor="#FFDF95" | | |||
''' ''Cryptomonas paramaecium'' [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=2898]''' | |||
|} | |||
{| | |||
| height="10" bgcolor="#FFDF95" | | |||
''' ''Cryptomonas phaseolus'' [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=70450&lvl=3&keep=1&srchmode=1&unlock&lin=f]''' | |||
|} | |||
{| | |||
| height="10" bgcolor="#FFDF95" | | |||
''' ''Cryptomonas pyrenoidifera'' [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=233184]''' | |||
|} | |||
{| | |||
| height="10" bgcolor="#FFDF95" | | |||
''' ''Cryptomonas tetrapyrenoidosa'' [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=233185]''' | |||
|} | |||
==Description and Significance== | ==Description and Significance== | ||
Cryptomonas | ''Cryptomonas'' is a genus of phytoplankton that inhabit freshwater aquatic systems. The genus belongs to a larger collective of organisms called "cryptomonads", which consists of of biflagellate organisms. Cells tend to be rather large, varying in shape between ovoid and elliptical, averaging 40 um in size. Cryptomonads can be either protozoans (order Cryptomonadida) or alga (class Cryptophyceae). Cryptomonas phytoplankton function as primary producers in freshwater ecosystems and contribute to the foraging base of many heterotrophs, including macroinvertibrates and fishes. While species coexist in the same habitats, they follow sequential blooms and occupy different spatial niches within the water column, which lends evidence to solve the "paradox of the plankton". | ||
[[Image:Cryptomonas_phylogeny.png|frame|left| The phylogenetic tree for the genus ''Cryptomonas''. From [http://tolweb.org/Public/treeImages/Cryptomonas.png?x=440164009 Tree of Life Web Project]]] | |||
==Genome Structure== | ==Genome Structure== | ||
Originally this genus existed as three seperate genuses: ''Cryptomonas'', ''Campylomonas'', and ''Chilomonas''. However, after the analysis of two nuclear ribosomal DNA regions and and a nucleomorph ribosomal gene, ''Campylomonas'' and ''Chilomonas'' were found to be the result of life history-dependent dimorphism in the ''Cryptomonas'' genus, and where therefore reclassified. [1] | |||
Species within the genus ''Cryptomonas'' have 4 genomes within the cell: the nucleus, the nucleomorph, the plastid, and the mitochondrial genomes. The plastid genome is circular and contains 118 kilobase pairs. This genome is the direct result of secondary endosymbiosis via a viral phage from a red alga, which accounts for the presence of red pigments within Cryptomonas species while other related genuses lack these pigments.[2] | |||
==Cell Structure, Metabolism and Life Cycle== | ==Cell Structure, Metabolism and Life Cycle== | ||
Cryptomonas are unique in their possession of two flagella, extending off either side of the cell. These structures are anchored within the cell by a unique configuration of four microtubular roots that work in cooperation with a striated fibrous root. The microtubular roots enter the anterior dorsal lobe of each Cryptomonas, while the striated fibrous root will be joined by five small microtubular roots surrounding the outside before anchoring. [5] | |||
To aide in movement, Cryptomonas has special hairs lining the length of their flagella. So-called Flimmer Hairs are slender extensions that occur in two rows from one flagellum, and one slightly smaller row from the other. They are capable of altering water movement surrounding the cell, thereby shifting the direction of movement of the cell itself. [6] | |||
Cryptomonas is a photolithotroph, performing photosynthesis for energy production while requiring inorganic compounds (often water) to act as electron donors. These cells also contribute greatly to oxygenic carbon fixation, rendering them crucial to carbon fluxes of fresh water environments. This fixation seems to be inversely proportional to sulfide levels, but proportional to polyglucose (forms of starch) and polyprotein production levels; polyglucose levels accumulated throughout the day are depleted at night. [9] | |||
Cryptomonas replication is dominant during the early summer bloom of fresh water species. Replication of these cells proceeds through mitosis, similar to many species. Chromatin within the cell condenses in prophase to form an equatorial plate, 0.7 um in thickness. Anaphase will allow division of the plate into two approximately equal but separate chromosomes, which will encircle within themselves during telophase; cytokinesis will proceed from here. The entire replication process only requires ten minutes. [7] | |||
==Ecology== | |||
''Cryptomonas'' species are almost always found in freshwater, and function like Diatoms, a related class of algae, in that they are large, slow-growing, and are often nutrient limited in their densities as opposed to predation pressure. Many varieties of Cryptomonas undergo blooms during winter when the limnion is covered in ice, whereas most other genera of phytoplankton undergo blooms in the summer months. [10] Their prevalence seems contrary to their slow generation times in comparison to other phytoplankton, as well as the fact that they are the primary food source of countless species of zooplankton, macroinvertibrates and fish. Indeed, there has been a study on a prevalent species of zooplantkon (''Ceriodaphnia quadrangula'') that demonstrated this species feeds exclusively on ''Cryptomonas erosa'', despite being saturated by larger and more nutrient-rich diatoms. [3] | |||
[[Image:Lake_clio.gif|frame|right| Lake Clio, Spain. This location is the site of the Pedros et al study on the layering of ''C. phaseolus'' From [http://starcentral.mbl.edu/msr/rawdata/viewable/cryptomonas12_tkw.jpg micro*scope]]] | |||
Further studies have demonstrated that ''Cryptomonas'' phytoplankton may actually migrate between layers of the water column to alternate between depths optimum for photosynthesis and bacteriograzing, and zones that deter mobile organisms that predate on the phytoplankon. Movement between these "feeding" and "refuge" zones was first observed in a study by Gasol, J. M. et al. Gasol observed that populations of C. phaseolus concentrated at the oxygen/sulfide interface within the lake, despite the fact that not only did sulfide inhibit the carbon fixation of these phytoplankton, but also that light levels were far below those that allowed for optimum growth rates. There was also a distinct lack of bacteriograzing, which is a behavior all cryptomonads demonstrate. Yet the population seemed to thrive with little competition from other species and genera of phytoplankton. [4] | |||
Later studies revealed that ''C. phaseolus'' moves through the water column in a diurnal cycle. During the day, the phytoplankton are found in the photic zone of their native lake, bacteriograzing and photosynthesizing in the presence of predators. At night, C. phaseolus re-accumulated at the hypolimnion where sulfide levels were high and oxygen levels were low: it was enough to deter large zooplankton from following, which reduced losses from predation by 38%. Indeed, grazing pressure only removed 5-25% of the total biomass of ''C. phaseolus'' daily, all of which the phytoplankton regenerated. [8] | |||
==References== | ==References== | ||
[ | [1] [http://www.biomedcentral.com/1471-2148/5/56 Hoef-Emden, K., Melkonian, M. "Revision of the Genus ''Cryptomonas'' (Cryptophyceae): a Combination of Molecular Phylogeny and Morphology Provides Insights into a Long-Hidden Dimorphism". ''Protist''. 2008. Volume 159:3, p. 507] | ||
[2] [http://www.biomedcentral.com/1471-2148/5/56 Hoef-Emden, K., Tran, H., Melkonian, M. "Lineage-specific variations of congruent evolution among DNA sequences from three genomes, and relaxed selective constraints on rbcL in ''Cryptomonas'' (Cryptophyceae)". ''BMC Evolutionary Biology''. 2005. Volume 5. p. 103-112.] | |||
[3] [http://www.springerlink.com/content/n654552l99h18506/ Gladyshev, M. I., Temerova, T. A., Dubovskaya, O. P., Kolmakov, V. I., Ivanova, E. A. "Selective grazing on ''Cryptomonas'' by ''Ceriodaphnia quadrangula'' fed a natural phytoplankton assemblage". ''Aquatic Ecology''. 1999. Volume 33:4. p. 347-353.] | |||
[4] [http://www.jstor.org/pss/4218893 Gasol, J. M., Garcia-Cantizano, J., Massana, R., Guerrero, R., Pedros-Alio, C. "Physiological ecology of a metalimnetic ''Cryptomonas'' population: relationships to light, sulfide, and nutrients". ''Journal of Plankton Research''. 1993. Volume 15:3. p. 255-275.] | |||
[5] [http://www3.interscience.wiley.com/journal/119525640/abstract?CRETRY=1&SRETRY=0 Roberts, Keith R. "Structure and Significance of the Cryptomonad Flagellar Apparatus". Journal of Phycology. 2004. Volume: 20:4. p. 590-599.] | |||
[6] [http://jcs.biologists.org/cgi/reprint/7/2/445.pdf Greenwood, A.D., et al. "The Origin of Flimmer in Cryptomonas". Cell Science (England). 1970. Volume 7. p. 445-461.] | |||
[7] [http://jcs.biologists.org/cgi/reprint/31/1/53.pdf Heath, I. Brent. and Oakley, Berl. "The Arrangement of Microtubules in Serially Sectioned Spindles of the Cryptomonas". Cell Science (England). 1978. Volume 31. p. 53-70.] | |||
[8] [http://www.icm.csic.es/bio/personal/mlatasa/Publications/Pedros%20et%20al%201995%20JPR.pdf Pedros-Alios, C., Massana, R., Latassa, M., Garcia-Cantizano, J., Gasol, J. M. "Predation by ciliares on a metalimnetic ''Cryptomonas'' population: feeding rates, impact and effects of vertical migration". ''Journal of Plankton Research''. 1995. Volume 17:11. p. 2131-2154.] | |||
[9] [http://plankt.oxfordjournals.org/cgi/reprint/15/3/255.pdf Garcia-Cantizano, Josefina, et al. "Physiological ecology of a metalimnetic Cryptomonas population". Journal of Plankton Research. 1993. Volume 15 no. 3. p. 255-275.] | |||
[10] Klaveness, D. "Ecology of the Cryptomonadida - a first review." ''Growth and Reproductive Strategies of Freshwater Phytoplankton'' pp. 105-133. Cambridge University Press, New York. | |||
==Author== | ==Author== | ||
Page authored by | Page authored by Allison Cutter and Alexandra David, students of [http://www.kbs.msu.edu/faculty/lennon/ Prof. Jay Lennon] at Michigan State University. | ||
[[Category:Pages edited by students of Jay Lennon at Michigan State University]] | [[Category:Pages edited by students of Jay Lennon at Michigan State University]] | ||
Latest revision as of 18:54, 25 August 2010

Classification
Eukaryota; Cryptophyta; Cryptomonadales; Cryptomonadaceae; Cryptomonas
Species
|
Cryptomonas borealis [1] |
|
Cryptomonas commutata [2] |
|
Cryptomonas curvata [3] |
|
Cryptomonas erosa [4] |
|
Cryptomonas gyropyrenoidosa [5] |
|
Cryptomonas lundii [6] |
|
Cryptomonas loricata [7] |
|
Cryptomonas marssonii [8] |
|
Cryptomonas obovoidea [9] |
|
Cryptomonas ovata [10] |
|
Cryptomonas paramaecium [11] |
|
Cryptomonas phaseolus [12] |
|
Cryptomonas pyrenoidifera [13] |
|
Cryptomonas tetrapyrenoidosa [14] |
Description and Significance
Cryptomonas is a genus of phytoplankton that inhabit freshwater aquatic systems. The genus belongs to a larger collective of organisms called "cryptomonads", which consists of of biflagellate organisms. Cells tend to be rather large, varying in shape between ovoid and elliptical, averaging 40 um in size. Cryptomonads can be either protozoans (order Cryptomonadida) or alga (class Cryptophyceae). Cryptomonas phytoplankton function as primary producers in freshwater ecosystems and contribute to the foraging base of many heterotrophs, including macroinvertibrates and fishes. While species coexist in the same habitats, they follow sequential blooms and occupy different spatial niches within the water column, which lends evidence to solve the "paradox of the plankton".
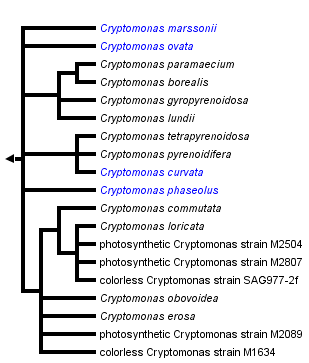
Genome Structure
Originally this genus existed as three seperate genuses: Cryptomonas, Campylomonas, and Chilomonas. However, after the analysis of two nuclear ribosomal DNA regions and and a nucleomorph ribosomal gene, Campylomonas and Chilomonas were found to be the result of life history-dependent dimorphism in the Cryptomonas genus, and where therefore reclassified. [1]
Species within the genus Cryptomonas have 4 genomes within the cell: the nucleus, the nucleomorph, the plastid, and the mitochondrial genomes. The plastid genome is circular and contains 118 kilobase pairs. This genome is the direct result of secondary endosymbiosis via a viral phage from a red alga, which accounts for the presence of red pigments within Cryptomonas species while other related genuses lack these pigments.[2]
Cell Structure, Metabolism and Life Cycle
Cryptomonas are unique in their possession of two flagella, extending off either side of the cell. These structures are anchored within the cell by a unique configuration of four microtubular roots that work in cooperation with a striated fibrous root. The microtubular roots enter the anterior dorsal lobe of each Cryptomonas, while the striated fibrous root will be joined by five small microtubular roots surrounding the outside before anchoring. [5]
To aide in movement, Cryptomonas has special hairs lining the length of their flagella. So-called Flimmer Hairs are slender extensions that occur in two rows from one flagellum, and one slightly smaller row from the other. They are capable of altering water movement surrounding the cell, thereby shifting the direction of movement of the cell itself. [6]
Cryptomonas is a photolithotroph, performing photosynthesis for energy production while requiring inorganic compounds (often water) to act as electron donors. These cells also contribute greatly to oxygenic carbon fixation, rendering them crucial to carbon fluxes of fresh water environments. This fixation seems to be inversely proportional to sulfide levels, but proportional to polyglucose (forms of starch) and polyprotein production levels; polyglucose levels accumulated throughout the day are depleted at night. [9]
Cryptomonas replication is dominant during the early summer bloom of fresh water species. Replication of these cells proceeds through mitosis, similar to many species. Chromatin within the cell condenses in prophase to form an equatorial plate, 0.7 um in thickness. Anaphase will allow division of the plate into two approximately equal but separate chromosomes, which will encircle within themselves during telophase; cytokinesis will proceed from here. The entire replication process only requires ten minutes. [7]
Ecology
Cryptomonas species are almost always found in freshwater, and function like Diatoms, a related class of algae, in that they are large, slow-growing, and are often nutrient limited in their densities as opposed to predation pressure. Many varieties of Cryptomonas undergo blooms during winter when the limnion is covered in ice, whereas most other genera of phytoplankton undergo blooms in the summer months. [10] Their prevalence seems contrary to their slow generation times in comparison to other phytoplankton, as well as the fact that they are the primary food source of countless species of zooplankton, macroinvertibrates and fish. Indeed, there has been a study on a prevalent species of zooplantkon (Ceriodaphnia quadrangula) that demonstrated this species feeds exclusively on Cryptomonas erosa, despite being saturated by larger and more nutrient-rich diatoms. [3]

Further studies have demonstrated that Cryptomonas phytoplankton may actually migrate between layers of the water column to alternate between depths optimum for photosynthesis and bacteriograzing, and zones that deter mobile organisms that predate on the phytoplankon. Movement between these "feeding" and "refuge" zones was first observed in a study by Gasol, J. M. et al. Gasol observed that populations of C. phaseolus concentrated at the oxygen/sulfide interface within the lake, despite the fact that not only did sulfide inhibit the carbon fixation of these phytoplankton, but also that light levels were far below those that allowed for optimum growth rates. There was also a distinct lack of bacteriograzing, which is a behavior all cryptomonads demonstrate. Yet the population seemed to thrive with little competition from other species and genera of phytoplankton. [4]
Later studies revealed that C. phaseolus moves through the water column in a diurnal cycle. During the day, the phytoplankton are found in the photic zone of their native lake, bacteriograzing and photosynthesizing in the presence of predators. At night, C. phaseolus re-accumulated at the hypolimnion where sulfide levels were high and oxygen levels were low: it was enough to deter large zooplankton from following, which reduced losses from predation by 38%. Indeed, grazing pressure only removed 5-25% of the total biomass of C. phaseolus daily, all of which the phytoplankton regenerated. [8]
References
[10] Klaveness, D. "Ecology of the Cryptomonadida - a first review." Growth and Reproductive Strategies of Freshwater Phytoplankton pp. 105-133. Cambridge University Press, New York.
Author
Page authored by Allison Cutter and Alexandra David, students of Prof. Jay Lennon at Michigan State University.
