Dietzia cinnamea: Difference between revisions
(Created page with "Environment Samples of this organism have been extracted from petroleum contaminated soil characterized in acidic sandy loam Cambisol soil in a protected habitat in Rio de Janei...") |
No edit summary |
||
| (65 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | |||
==Classification== | |||
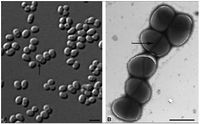
[[Image:D_cin_pic.jpg|thumb|Right|frame|200px| Figure 1.''D.cinnamea'' displaying coccoid to ovoid shape and colonies (Von der Weid, 2006).]] | |||
D. cinnamea is capable of degrading a range of petroleum hydrocarbons which can have beneficial environmental implications in today’s world. Other genera that have hydrocarbon degrading strains include Mycobacterium, Rhodococcus, and Dietzia. | Domain: Bacteria | ||
The strain P4 was found to | |||
Phylum: Actinobacteria | |||
Class: Actinobacteria | |||
Order: Actinomycetales | |||
Sub-order: Corynebacterineae | |||
Family: Dietziaceae | |||
Genus: ''Dietzia'' | |||
'''NCBI Taxonomy ID:[http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=910954&lvl=3&lin=f&keep=1&srchmode=1&unlock]''' | |||
Species: ''Dietzia cinnamea'' | |||
==Description and Significance== | |||
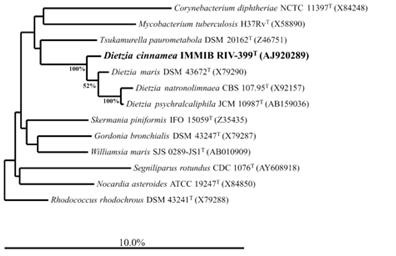
[[Image:D_cin_newtable.jpg|400px|right|frame| Figure 2. Neighbour-joining tree showing the position of strain IMMIB RIV-399T (=DSM 44904T=CCUG 50875T) within the radiation of the mycolic acid-containing taxa. Thetree was based on a comparison of sequences that were at least 90 % complete (with regard to E. coli sequence). Bar, 10·0 % sequence divergence (Yassin, 2006).]] | |||
Samples of this microbe have been found in nature twice. The first was from a perianal swab from a patient with a bone marrow transplant (Yassin, 2006). The second, named P4, was extracted from petroleum contaminated soil characterized in acidic sandy loam Cambisol soil in a protected habitat in Rio de Janeiro state, Brazil (Von der Weid, 2006). | |||
Multiple strains of Dietzia have been found in soil, deep sea sediment, and soda lakes (Gerday & Glansdorff, 2007).Dietzia cinnamea is rod shaped in the medical swab while the P4 strain from the soil samples produces a coccoid shape (Figure 1). The organism is approximately 1.4 micrometers long, forms smooth, yellow to orange colonies on agar plates once isolated and is single or arranges in small connected colonies (Von der Weid, 2006). It is gram positive and has a high G+C content, meaning a high number of Guanine and Cytosine linkages in its DNA (Von der Weid, 2006). It displays snapping division, which is the arrangement of cells in a palisade or angular manor after binary fission (Prescott, 2002). This is a characteristic of the genera Arthrobacter and Corynebacterium. | |||
D. cinnamea is capable of degrading a range of petroleum hydrocarbons which can have beneficial environmental implications in today’s world. Other genera that have hydrocarbon degrading strains include Mycobacterium, Rhodococcus, and Dietzia. The strain P4 is able to degrade a range of n-alkanes (C11-C36), pristane, and phytane and is able to grow in the presence of carbazole, quinoline, naphthalene, toluene, gasoline, and diesel (Von der Weid, 2006). | |||
==Genome Structure== | |||
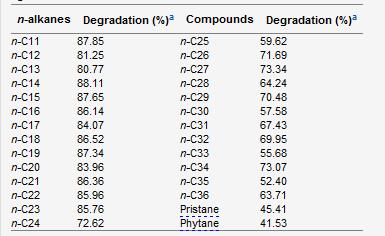
[[Image:Carbon Sources.jpg|right|frame|Figure 3. nd = not determined. ++ is very good growth. + is good growth. - is no growth observed. Different carbon sources tested for metabolic use by ''D.cinnamea'' P4 (Von der Weid, 2006)]] | |||
The full DNA code for ''D.cinnamea'' has been partially sequenced and contains 3,555,295 bp (NCBI Nucleotide):[http://www.ncbi.nlm.nih.gov/sites/entrez?db=nuccore&cmd=DetailsSearch&term=dietzia+cinnamea&save_search=true]. | |||
''D.cinnamea'' has a GC content of 72.3% (Von der Weid, 2006). | |||
DNA of the P4 strain was sequenced with a 16S rDNA and it was found to have a 99.8% similarity to ''D.cinnamea'' that was found in the previous bone marrow transplant swab. When testing DNA-DNA similarities, the strains were 93.3% homologous with the transplant swab. A 70% threshold of DNA-DNA similarity is required to include it in the species. This confirms that P4 is part of the species ''D.cinnamea''. This species has a 30-50% genome similarity with the other species of ''Dietzia'' (''D.maris'' 52.4%, ''D.natronolimnaea''' 56.3%, ''D.psychraicliphila'' 37.8%)(Figure 2)(Von der Weid, 2006). | |||
==Cell Structure, Metabolism, and Life Cycle== | |||
''D.cinnamea'' is gram positive and ranges from rod to coccoid shaped. Average cell size is approximately 1.4 micrometers. | |||
D.cinnamea is an aerobic organotroph that can utilize acetate, D-glucose, maltose, and 1,2 propandediol as a carbon source (Yassin, 2006). P4 uses maltose as a carbon source in addition to petroleum hydrocarbons including n-alkanes, toluene, diesel, gasoline, naphthalene, quinoline, and carbazole (Figure 3)(Von der Weid, 2006). Successful growth was achieved when the petroleum hydrocarbons were used with mineral liquid media. P4 was also able to grow on Arabian light oil and Marlin oil without additional carbon or nitrogen sources required (Von der Weid, 2006). ''D.cinnamea'' is therefore a useful microbe in bioremediation given its potential break down of petroleum based hydrocarbons. Specific mechanisms of hydrocarbon bioremediation is not fully understood currently (Iwaki, 2008). Metabolic activities reduce nitrate to nitrite (Von der Weid, 2006). | |||
[[Image:Degradation.jpg|frame|Figure 4. The reduction capabilities of ''D.cinnamea'' P4 on various n-alkanes and pristane and phytane(Von der Weid, 2006)]] | |||
==Ecology and Pathogenesis== | |||
The genera ''Dietzia'' has been found in multiple habitats including soil, deep sea sediment, soda lakes, marine aquatic life, and have been classified as a pathogen to humans. ''D.cinnamea'' has been extracted from the tropical soils of Brazil but could thrive in other related environments such as the genera does. The habitat in which P4 was first found was an acidic, sandy loam. Cultured samples grew best in pH ranging from 6 to 10 and optimally at 9. Growth can occur within a temperature of 30-45 degrees C with optimal growth at 32. These lab parameters can be associated with the habitat in which it was originally found. ''D.cinnamea'' is able to grow in the presence or absence of 10% NaCl making showing it is slightly halotolerant (Von der Weid, 2006). | |||
First discovery of the species was from a bone marrow transplant patient therefore making it a possible human pathogen, however further research is needed. Given its diverse metabolic abilities, ''D.cinnamea'' has potential to serve a role in bioremediation. P4 was the first representative of ''D.cinnamea'' with this capability. P4 is capable of degrading multiple n-alkanes (C11-C36), pristanes, and phytanes as hydrocarbon sources (Figure 4). Specifically, C11-C23 were all degraded by at least 80% by ''D.cinnamea'' (Figure 4). Samples in lab thrived in petroleum fed parameters indicating this microbe would be effective to use in bioremediation. Further research needs to be conducted in order to know if this microbe works ''in situ'' which would aid efforts to naturally clean petroleum polluted areas (Von der Weid, 2006). | |||
==References== | |||
Gerday, C., & Glansdorff, N. (2007). Physiology and biochemistry of extremophiles. Washington, D.C.: American Society for Mircrobiology Press. | |||
Iwaki, H., Nakai, E., Nakamura, S., Hasegawa, Y. Isolation and Characterization of Cyclohexylacetic Acid-degrading Bacteria. Current Microbiology. 2008. 57:107-110. | |||
"Dietzia cinnamea P4, whole genome shotgun sequence". Entrez Genome Project. NCBI. Accessed April 23, 2011. | |||
Prescott, L., Klein, D., & Harley, J. (2002). Microbiology. Retrieved from Online Learning Center: http://highered.mcgraw-hill.com/sites/0072320419/student_view0/glossary_s-z.html | |||
Von Der Weid, I., Marques, J. M., Cunha, C. D., Lippi, R. K., Dos Santos, S. C. C., Rosado, A. S., Lins, U., et al. (2007). Identification and biodegradation potential of a novel strain of Dietzia cinnamea isolated from a petroleum-contaminated tropical soil. Systematic and applied microbiology, 30(4), 331-339. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/1717450 | |||
Yassin, A., Hupfer, H., & Schaal, K. (2006). Dietzia cinnamea sp.nov., a novel species isolated from a perianal swab of a patient with a bone marrow transplant. International Journal of Systematic and Evolutionary Microbiology , 641-645. | |||
==Author== | |||
Page authored by Jennifer Jury and Elizabeth Karinen, students of [http://www.kbs.msu.edu/faculty/lennon/ Prof. Jay Lennon] at Michigan State University. | |||
[[Category:Pages edited by students of Jay Lennon at Michigan State University]] | |||
Latest revision as of 19:33, 1 September 2011
Classification
Domain: Bacteria
Phylum: Actinobacteria
Class: Actinobacteria
Order: Actinomycetales
Sub-order: Corynebacterineae
Family: Dietziaceae
Genus: Dietzia
NCBI Taxonomy ID:[1]
Species: Dietzia cinnamea
Description and Significance
Samples of this microbe have been found in nature twice. The first was from a perianal swab from a patient with a bone marrow transplant (Yassin, 2006). The second, named P4, was extracted from petroleum contaminated soil characterized in acidic sandy loam Cambisol soil in a protected habitat in Rio de Janeiro state, Brazil (Von der Weid, 2006). Multiple strains of Dietzia have been found in soil, deep sea sediment, and soda lakes (Gerday & Glansdorff, 2007).Dietzia cinnamea is rod shaped in the medical swab while the P4 strain from the soil samples produces a coccoid shape (Figure 1). The organism is approximately 1.4 micrometers long, forms smooth, yellow to orange colonies on agar plates once isolated and is single or arranges in small connected colonies (Von der Weid, 2006). It is gram positive and has a high G+C content, meaning a high number of Guanine and Cytosine linkages in its DNA (Von der Weid, 2006). It displays snapping division, which is the arrangement of cells in a palisade or angular manor after binary fission (Prescott, 2002). This is a characteristic of the genera Arthrobacter and Corynebacterium.
D. cinnamea is capable of degrading a range of petroleum hydrocarbons which can have beneficial environmental implications in today’s world. Other genera that have hydrocarbon degrading strains include Mycobacterium, Rhodococcus, and Dietzia. The strain P4 is able to degrade a range of n-alkanes (C11-C36), pristane, and phytane and is able to grow in the presence of carbazole, quinoline, naphthalene, toluene, gasoline, and diesel (Von der Weid, 2006).
Genome Structure
The full DNA code for D.cinnamea has been partially sequenced and contains 3,555,295 bp (NCBI Nucleotide):[2].
D.cinnamea has a GC content of 72.3% (Von der Weid, 2006).
DNA of the P4 strain was sequenced with a 16S rDNA and it was found to have a 99.8% similarity to D.cinnamea that was found in the previous bone marrow transplant swab. When testing DNA-DNA similarities, the strains were 93.3% homologous with the transplant swab. A 70% threshold of DNA-DNA similarity is required to include it in the species. This confirms that P4 is part of the species D.cinnamea. This species has a 30-50% genome similarity with the other species of Dietzia (D.maris 52.4%, D.natronolimnaea' 56.3%, D.psychraicliphila 37.8%)(Figure 2)(Von der Weid, 2006).
Cell Structure, Metabolism, and Life Cycle
D.cinnamea is gram positive and ranges from rod to coccoid shaped. Average cell size is approximately 1.4 micrometers.
D.cinnamea is an aerobic organotroph that can utilize acetate, D-glucose, maltose, and 1,2 propandediol as a carbon source (Yassin, 2006). P4 uses maltose as a carbon source in addition to petroleum hydrocarbons including n-alkanes, toluene, diesel, gasoline, naphthalene, quinoline, and carbazole (Figure 3)(Von der Weid, 2006). Successful growth was achieved when the petroleum hydrocarbons were used with mineral liquid media. P4 was also able to grow on Arabian light oil and Marlin oil without additional carbon or nitrogen sources required (Von der Weid, 2006). D.cinnamea is therefore a useful microbe in bioremediation given its potential break down of petroleum based hydrocarbons. Specific mechanisms of hydrocarbon bioremediation is not fully understood currently (Iwaki, 2008). Metabolic activities reduce nitrate to nitrite (Von der Weid, 2006).
Ecology and Pathogenesis
The genera Dietzia has been found in multiple habitats including soil, deep sea sediment, soda lakes, marine aquatic life, and have been classified as a pathogen to humans. D.cinnamea has been extracted from the tropical soils of Brazil but could thrive in other related environments such as the genera does. The habitat in which P4 was first found was an acidic, sandy loam. Cultured samples grew best in pH ranging from 6 to 10 and optimally at 9. Growth can occur within a temperature of 30-45 degrees C with optimal growth at 32. These lab parameters can be associated with the habitat in which it was originally found. D.cinnamea is able to grow in the presence or absence of 10% NaCl making showing it is slightly halotolerant (Von der Weid, 2006).
First discovery of the species was from a bone marrow transplant patient therefore making it a possible human pathogen, however further research is needed. Given its diverse metabolic abilities, D.cinnamea has potential to serve a role in bioremediation. P4 was the first representative of D.cinnamea with this capability. P4 is capable of degrading multiple n-alkanes (C11-C36), pristanes, and phytanes as hydrocarbon sources (Figure 4). Specifically, C11-C23 were all degraded by at least 80% by D.cinnamea (Figure 4). Samples in lab thrived in petroleum fed parameters indicating this microbe would be effective to use in bioremediation. Further research needs to be conducted in order to know if this microbe works in situ which would aid efforts to naturally clean petroleum polluted areas (Von der Weid, 2006).
References
Gerday, C., & Glansdorff, N. (2007). Physiology and biochemistry of extremophiles. Washington, D.C.: American Society for Mircrobiology Press.
Iwaki, H., Nakai, E., Nakamura, S., Hasegawa, Y. Isolation and Characterization of Cyclohexylacetic Acid-degrading Bacteria. Current Microbiology. 2008. 57:107-110.
"Dietzia cinnamea P4, whole genome shotgun sequence". Entrez Genome Project. NCBI. Accessed April 23, 2011.
Prescott, L., Klein, D., & Harley, J. (2002). Microbiology. Retrieved from Online Learning Center: http://highered.mcgraw-hill.com/sites/0072320419/student_view0/glossary_s-z.html
Von Der Weid, I., Marques, J. M., Cunha, C. D., Lippi, R. K., Dos Santos, S. C. C., Rosado, A. S., Lins, U., et al. (2007). Identification and biodegradation potential of a novel strain of Dietzia cinnamea isolated from a petroleum-contaminated tropical soil. Systematic and applied microbiology, 30(4), 331-339. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/1717450
Yassin, A., Hupfer, H., & Schaal, K. (2006). Dietzia cinnamea sp.nov., a novel species isolated from a perianal swab of a patient with a bone marrow transplant. International Journal of Systematic and Evolutionary Microbiology , 641-645.
Author
Page authored by Jennifer Jury and Elizabeth Karinen, students of Prof. Jay Lennon at Michigan State University.