Vibrio harveyi: Difference between revisions
No edit summary |
|||
| (126 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
{{Curated}} | |||
{{Biorealm Genus}} | {{Biorealm Genus}} | ||
| Line 17: | Line 18: | ||
===Species=== | ===Species=== | ||
'''''Vibrio harveyi''''' | |||
''' | |||
''Vibrio | Synonyms: ''Lucibacterium harveyi'', ''Beneckea harveyi'', ''Achromobacter harveyi'', ''Pseudomonas harveyi'', ''Photobacterium harveyi'', ''Vibrio carchariae'', ''Vibrio trachuri''; (Thompson, 2002) | ||
==Description and significance== | ==Description and significance== | ||
: | [[Image:Vh1.JPG|thumb|Courtesy of Bonnie Bassler Lab]] | ||
''V. harveyi'' is a natural inhabitant of seawater, though it has been documented mainly in the tropics. Unlike its close relative [[''Vibrio fischeri'']] it is free-living and does not form symbiotic relationships, but is a common pathogen to many marine organisms. In applied science ''Vibrio harveyi'' is an integral part of studying inter- and intra-bacterial communication, quorum sensing, as well as bioluminescence. Sequencing of the genome will undoubtedly contribute to further understanding of how bacteria sense the world around them and then make decisions based on their environment. ''V. harveyi'' is curved rod shaped, Gram-negative, and monotrichous (mobilized by a single polar flagellum). | |||
==Genome structure== | |||
:The Microbial Sequencing Center of the Institute for Genomic Research is currently doing shotgun seguence research for the HY01 strain of ''Vibrio harveyi''. This research is in conjunction with a project to identify the genome sequence for 17 different Vibrio species enabling extensive comprehension of the Vibrio genus. | :The Microbial Sequencing Center of the Institute for Genomic Research is currently doing shotgun seguence research for the HY01 strain of ''Vibrio harveyi''. This research is in conjunction with a project to identify the genome sequence for 17 different Vibrio species enabling extensive comprehension of the Vibrio genus. | ||
:The Genome Sequencing Center of the Washington University School of Medicine is also working on sequencing the genome of ''V. harveyi.'' The center believes sequencing the ''V. harveyi'' genome is extremely important especially because of its unique communication skills, and the scientific community has agreed (read on). The website hosts a research status page. [http://genome.wustl.edu/genome.cgi?GENOME=Vibrio%20harveyi&SECTION=research] | |||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
''V. harveyi'' is Gram-negative, therefore it has a cell wall that consists of two membranes: an outer membrane made up of lipopolysaccharides and an inner cytoplasmic membrane. In between these is a periplasmic space housing a peptidoglycan layer. | |||
''Vibrio harveyi'' is a facultative anaerobe, meaning it can swap between aerobic respiration of oxygen and fermentation in order to produce ATP. This characteristic helps ''V. harveyi'' survive in low oxygen concentrations, if fermentable material is present. ''V. harveyi'' has the ability of bioluminescence, which is controlled by the regulatory gene luxR as well as population density-sensing (quorum sensing) autoinducer signaling molecules. Expression occurs when the enzyme luciferace oxidizes an organic compound releasing free energy in the form of light. (Showalter, 1990) | |||
''Vibrio harveyi'' is known to regulate gene expression based on its population density. This is known as "Quorum-Sensing". This cell-to-cell communication ability is due to the release of autoinducers as a function of population in a given area and can occur between different bacterial species. Some examples of regulated activities based on quorum sensing include virulence, antibiotic production, biofilm formation, symbiosis, motility, and luminescence. ''V. harveyi'' actually produces two different autoinducer signals; one for intraspecies communication (denoted AI-1) and one for interspecies communication (AI-2). (Henke, 2004) | |||
==Ecology== | ==Ecology== | ||
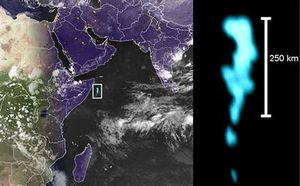
[[Image:Milkysea_miller.jpg|thumb|Courtesy of S. Miller, Haddock, Elvidge, and Lee]] | |||
The interspecial communication skills of ''Vibrio harveyi'' have major effects on the way the bacteria interact with the environment, whether it is luminescing, becoming virulent, or excreting nucleases for defense. Even though ''V. harveyi'' does not form symbiotic relationships, there is one well documented example of a relationship with another microorganism causing a phenomenon known as the Milky Sea effect. This occurs when large areas of the ocean are filled with luminescing "''V. harveyi'' living in association with colonies of the microalga ''Phaeocystis''." (Miller, 2005) | |||
==Pathology== | ==Pathology== | ||
''Vibrio harveyi'' is a pathogen of fish and invertebrates, including sharks, seabass, seahorses, lobster, and shrimp. Its pathogenicity depends on the concentration of ''V. harveyi'' cells at a given time. Diseases caused by ''V. harveyi'' include eye-lesions, gastro-enteritis, vasculitis, and luminous vibriosis. Luminous vibriosis is a leading cause of death among comercially farmed shrimp and other aquaculture. The infection, by ''V. harveyi'', enters through the mouth and forms plaques, then spreads to the inards and the appendages. Loss of limb function and appendage degradation has been documented. Contamination can spread all the way to egg and larval tanks, thus causing an even bigger problem for shrimp farmers. Luminous vibriosis has been documented in many other crustaceans all of which glow in the dark when infected. Mortality occurs when penaeid shrimp is exposed to a concentraion of ''V. harveyi'' at 102 cells/milliliter. (Lightner, 1993) | |||
''V. harveyi'' is non pathogenic to humans, making it an ideal candidate for studying the Vibrio genus. | |||
==Application to Biotechnology== | ==Application to Biotechnology== | ||
The interspecial autoinducer AI-2, produced by ''V. harveyi'' could be a significant molecule in biotechnology. Analysis of the autoinducer could lead to developement of synthetic inhibitors of the communication pathway. This interference has the possibility to disrupt virulence factors, thus leading to novel antimicrobials. | |||
::"AI-2 itself could well function as a broad-spectrum antibiotic that disrupts quorum-sensing systems in a wide variety of Gram-negative and Gram-positive pathogens." (Federle, 2003) | |||
==Current Research== | ==Current Research== | ||
Most of the current research on ''Vibrio harveyi'' is being done at the Bonnie L. Bassler Laboratory at Princeton. They are studying the effect of AI-1 and AI-2 signaling on virulence in ''Vibrio cholerae''. Recent research at the lab uncovered the regulatory mechanism for quorum sensing mode in these bacteria. | |||
The Bassler Lab also focuses and the development of molecules that are similar to AI-2 to be used as antimicrobials on bacteria that utilize the AI-2 system for virulence (such as cholera). | |||
''V. harveyi'' is continuosly studied by various aquaculture protection agencies in the Bahamas, Asia, and the Philippines, focusing on its pathogenic quality and its effect on the worldwide supply of seafood. [http://www.cfsan.fda.gov/~ear/sea-glow.html] | |||
As mentioned earlier, the sequencing of the ''Vibrio harveyi'' genome is in progress. | |||
==References== | ==References== | ||
Edited by Ryan Kendall of [mailto:ralarsen@ucsd.edu Rachel Larsen]'s Bacteriology class Spring 2007 | 1. Austin B, and Zhang XH. Vibrio harveyi: a significant pathogen of marine vertebrates and invertebrates. Lett Appl Microbiol. 2006 Aug;43(2):119-24. | ||
2. Diggles, BK. Luminous vibriosis in rock lobster Jasus verreauxi (Decapoda: Palinuridae) phyllosoma larvae associated with infection by Vibrio harveyi. Diseases of Aquatic Organisms. 2000. 43(2):127-37 | |||
3. Federle, Michael J. and Bonnie L. Bassler. Interspecies communication in bacteria. Journal of Clinical Investigation. 2003. 112:1291-1299 | |||
4. Gauger, EJ. 16S ribosomal DNA sequencing confirms the synonymy of Vibrio harveyi and V. carchariae. Diseases of aquatic organisms. 2002 Nov 7;52(1):39-46. | |||
5. Henke, Jennifer M. and Bonnie L. Bassler. Three Parallel Quorum-Sensing Systems Regulate Gene Expression in Vibrio harveyi. J. Bacteriol. 2004 186: 6902-6914. | |||
6. Iwamoto, Y. Vibrio trachuri sp. nov., a new species isolated from diseased Japanese horse mackerel. Microbiol. Immunol. 1995. 39(11):831-37 | |||
7. Lightner, D. V. Diseases of cultured penaeid shrimp. CRC Handbook of Mariculture, Crustacean Aquaculture. 1993 pp. 393–486. | |||
8. Miller, Melissa B. and Bonnie L. Bassler. Quorum sensing in bacteria. Annual Review of Microbiology. 2001. 55:165-199 | |||
9. Miller, S.D., S.H.D. Haddock, C.D. Elvidge, T.F. Lee. Detection of a bioluminescent milky sea from space. Proc. National Acadamy Science 2005. 102:14181-14184 | |||
10. Nealson, K.H. and J.W. Hastings. Quorum sensing on a global scale: massive numbers of bioluminescent bacteria make milky seas Applied and Environmental Microbiology. 2006. 72:2295-2297 | |||
11. Showalter, R.E., Martin, M.O., and Silverman, M.R. Cloning and nucleotide sequence of luxR, a regulatory gene controlling bioluminescence in Vibrio harveyi. J. Bacteriol. 1990. 172: 2946–2954. | |||
12. Thompson, FL. Vibrio trachuri Iwamoto et al. 1995 is a junior synonym of Vibrio harveyi (Johnson and Shunk 1936) Baumann et al. 1981. Int J Syst Evol Microbiol. 2002 May;52(Pt 3):973-6 | |||
Edited by Ryan Kendall of [mailto:ralarsen@ucsd.edu Rachel Larsen] and Kit Pogliano's Bacteriology class Spring 2007 | |||
Latest revision as of 15:41, 1 July 2011
A Microbial Biorealm page on the genus Vibrio harveyi
Classification
Higher Order Taxa
- Kingdom: Bacteria
- Phylum: Proteobacteria
- Class: Gammaproteobacteria
- Order: Vibrionales
- Family: Vibrionaceae
Species
Vibrio harveyi
Synonyms: Lucibacterium harveyi, Beneckea harveyi, Achromobacter harveyi, Pseudomonas harveyi, Photobacterium harveyi, Vibrio carchariae, Vibrio trachuri; (Thompson, 2002)
Description and significance
V. harveyi is a natural inhabitant of seawater, though it has been documented mainly in the tropics. Unlike its close relative ''Vibrio fischeri'' it is free-living and does not form symbiotic relationships, but is a common pathogen to many marine organisms. In applied science Vibrio harveyi is an integral part of studying inter- and intra-bacterial communication, quorum sensing, as well as bioluminescence. Sequencing of the genome will undoubtedly contribute to further understanding of how bacteria sense the world around them and then make decisions based on their environment. V. harveyi is curved rod shaped, Gram-negative, and monotrichous (mobilized by a single polar flagellum).
Genome structure
- The Microbial Sequencing Center of the Institute for Genomic Research is currently doing shotgun seguence research for the HY01 strain of Vibrio harveyi. This research is in conjunction with a project to identify the genome sequence for 17 different Vibrio species enabling extensive comprehension of the Vibrio genus.
- The Genome Sequencing Center of the Washington University School of Medicine is also working on sequencing the genome of V. harveyi. The center believes sequencing the V. harveyi genome is extremely important especially because of its unique communication skills, and the scientific community has agreed (read on). The website hosts a research status page. [1]
Cell structure and metabolism
V. harveyi is Gram-negative, therefore it has a cell wall that consists of two membranes: an outer membrane made up of lipopolysaccharides and an inner cytoplasmic membrane. In between these is a periplasmic space housing a peptidoglycan layer. Vibrio harveyi is a facultative anaerobe, meaning it can swap between aerobic respiration of oxygen and fermentation in order to produce ATP. This characteristic helps V. harveyi survive in low oxygen concentrations, if fermentable material is present. V. harveyi has the ability of bioluminescence, which is controlled by the regulatory gene luxR as well as population density-sensing (quorum sensing) autoinducer signaling molecules. Expression occurs when the enzyme luciferace oxidizes an organic compound releasing free energy in the form of light. (Showalter, 1990)
Vibrio harveyi is known to regulate gene expression based on its population density. This is known as "Quorum-Sensing". This cell-to-cell communication ability is due to the release of autoinducers as a function of population in a given area and can occur between different bacterial species. Some examples of regulated activities based on quorum sensing include virulence, antibiotic production, biofilm formation, symbiosis, motility, and luminescence. V. harveyi actually produces two different autoinducer signals; one for intraspecies communication (denoted AI-1) and one for interspecies communication (AI-2). (Henke, 2004)
Ecology
The interspecial communication skills of Vibrio harveyi have major effects on the way the bacteria interact with the environment, whether it is luminescing, becoming virulent, or excreting nucleases for defense. Even though V. harveyi does not form symbiotic relationships, there is one well documented example of a relationship with another microorganism causing a phenomenon known as the Milky Sea effect. This occurs when large areas of the ocean are filled with luminescing "V. harveyi living in association with colonies of the microalga Phaeocystis." (Miller, 2005)
Pathology
Vibrio harveyi is a pathogen of fish and invertebrates, including sharks, seabass, seahorses, lobster, and shrimp. Its pathogenicity depends on the concentration of V. harveyi cells at a given time. Diseases caused by V. harveyi include eye-lesions, gastro-enteritis, vasculitis, and luminous vibriosis. Luminous vibriosis is a leading cause of death among comercially farmed shrimp and other aquaculture. The infection, by V. harveyi, enters through the mouth and forms plaques, then spreads to the inards and the appendages. Loss of limb function and appendage degradation has been documented. Contamination can spread all the way to egg and larval tanks, thus causing an even bigger problem for shrimp farmers. Luminous vibriosis has been documented in many other crustaceans all of which glow in the dark when infected. Mortality occurs when penaeid shrimp is exposed to a concentraion of V. harveyi at 102 cells/milliliter. (Lightner, 1993)
V. harveyi is non pathogenic to humans, making it an ideal candidate for studying the Vibrio genus.
Application to Biotechnology
The interspecial autoinducer AI-2, produced by V. harveyi could be a significant molecule in biotechnology. Analysis of the autoinducer could lead to developement of synthetic inhibitors of the communication pathway. This interference has the possibility to disrupt virulence factors, thus leading to novel antimicrobials.
- "AI-2 itself could well function as a broad-spectrum antibiotic that disrupts quorum-sensing systems in a wide variety of Gram-negative and Gram-positive pathogens." (Federle, 2003)
Current Research
Most of the current research on Vibrio harveyi is being done at the Bonnie L. Bassler Laboratory at Princeton. They are studying the effect of AI-1 and AI-2 signaling on virulence in Vibrio cholerae. Recent research at the lab uncovered the regulatory mechanism for quorum sensing mode in these bacteria.
The Bassler Lab also focuses and the development of molecules that are similar to AI-2 to be used as antimicrobials on bacteria that utilize the AI-2 system for virulence (such as cholera).
V. harveyi is continuosly studied by various aquaculture protection agencies in the Bahamas, Asia, and the Philippines, focusing on its pathogenic quality and its effect on the worldwide supply of seafood. [2]
As mentioned earlier, the sequencing of the Vibrio harveyi genome is in progress.
References
1. Austin B, and Zhang XH. Vibrio harveyi: a significant pathogen of marine vertebrates and invertebrates. Lett Appl Microbiol. 2006 Aug;43(2):119-24.
2. Diggles, BK. Luminous vibriosis in rock lobster Jasus verreauxi (Decapoda: Palinuridae) phyllosoma larvae associated with infection by Vibrio harveyi. Diseases of Aquatic Organisms. 2000. 43(2):127-37
3. Federle, Michael J. and Bonnie L. Bassler. Interspecies communication in bacteria. Journal of Clinical Investigation. 2003. 112:1291-1299
4. Gauger, EJ. 16S ribosomal DNA sequencing confirms the synonymy of Vibrio harveyi and V. carchariae. Diseases of aquatic organisms. 2002 Nov 7;52(1):39-46.
5. Henke, Jennifer M. and Bonnie L. Bassler. Three Parallel Quorum-Sensing Systems Regulate Gene Expression in Vibrio harveyi. J. Bacteriol. 2004 186: 6902-6914.
6. Iwamoto, Y. Vibrio trachuri sp. nov., a new species isolated from diseased Japanese horse mackerel. Microbiol. Immunol. 1995. 39(11):831-37
7. Lightner, D. V. Diseases of cultured penaeid shrimp. CRC Handbook of Mariculture, Crustacean Aquaculture. 1993 pp. 393–486.
8. Miller, Melissa B. and Bonnie L. Bassler. Quorum sensing in bacteria. Annual Review of Microbiology. 2001. 55:165-199
9. Miller, S.D., S.H.D. Haddock, C.D. Elvidge, T.F. Lee. Detection of a bioluminescent milky sea from space. Proc. National Acadamy Science 2005. 102:14181-14184
10. Nealson, K.H. and J.W. Hastings. Quorum sensing on a global scale: massive numbers of bioluminescent bacteria make milky seas Applied and Environmental Microbiology. 2006. 72:2295-2297
11. Showalter, R.E., Martin, M.O., and Silverman, M.R. Cloning and nucleotide sequence of luxR, a regulatory gene controlling bioluminescence in Vibrio harveyi. J. Bacteriol. 1990. 172: 2946–2954.
12. Thompson, FL. Vibrio trachuri Iwamoto et al. 1995 is a junior synonym of Vibrio harveyi (Johnson and Shunk 1936) Baumann et al. 1981. Int J Syst Evol Microbiol. 2002 May;52(Pt 3):973-6
Edited by Ryan Kendall of Rachel Larsen and Kit Pogliano's Bacteriology class Spring 2007