Gemmatimonas aurantiaca: Difference between revisions
| (76 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
==Classification== | ==Classification== | ||
===Higher order taxa=== | ===Higher order taxa=== | ||
Bacteria; Gemmatimonadetes; Gemmatimonadales; Gemmatimonadaceae; Gemmatimonas | Super Kingdom: Bacteria; Phylum: Gemmatimonadetes; Class: Gemmatimonadetes; Order: Gemmatimonadales; Family: Gemmatimonadaceae; Genus: ''Gemmatimonas'' | ||
(NCBI) | |||
===Species=== | ===Species=== | ||
Gemmatimonas aurantiaca T-27 | {| | ||
| height="10" bgcolor="#FFDF95" | | |||
'''NCBI: [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=379066&lvl=3&lin=f&keep=1&srchmode=1&unlock Taxonomy]''' | |||
|} | |||
Species: ''Gemmatimonas aurantiaca T-27T'' | |||
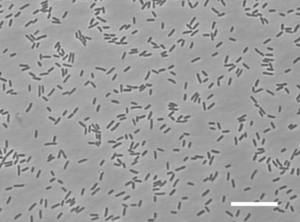
[[File:What_it_looks_like.gif|thumb|<i>G. aurantiaca T-27</i> under the microscope by Phase-contrast photomicrography [Zhang et al.]]] | |||
==Description and significance== | ==Description and significance== | ||
<i>G. aurantiaca | The phylum Gemmatimonadetes is a very recently described bacterial group whose members are widespread in nature, particularly in soil habitats. There is only one cultured representative of this phylum that is <i>Gemmatimonas aurantiaca</i>. [Joseph et al.] <i>G. aurantiaca T27T</i> is an aerobic rod-shaped Gram-negative bacterium that is non-sporulating. It is motile by the use of flagella and it was isolated by anaerobic-aerobic sequential batch reactor under enhanced biological phosphorus removal conditions for wastewater treatment. <i>G. aurantiaca T27T</i> is a very slow growing bacterium and is a polyphosphate-accummulating bacteria. It is able to grow not only by anaerobic respiration under anaerobic conditions but also by oxygen respiration under aerobic conditions. It is also orange in color, whether growing on a plate or in a liquid media. Due to <i>G. aurantiaca</i> having many distinct phylogenetic traits, it suggests that this bacterium will have many unique enzymes and metabolic pathways. Because of its uniqueness it was promised that this bacterium would lead to the discovery of new carotenoids. [Takaichi et al.] | ||
==Genome structure== | ==Genome structure== | ||
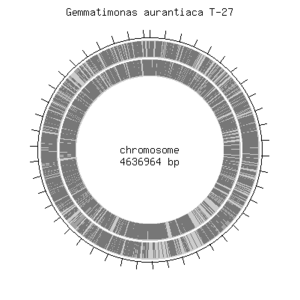
<i>Gemmatimonas aurantiaca T-27T</i> has a circular chromosome that consists of 4,636,964 base pairs of which majority, 64.28%, includes G/C pair. <i>G. aurantiaca</i> contains about 3,935 open reading frames, 48 tRNA genes and single rRNA operon were predicted. It also contains essential genes that were previously identified in model organisms such as <i>Escherichia coli</i> and <i>Bacillus subtilis</i>. This shows that their basic cellular system is not much different from other known microbes. <i>G. aurantiaca T-27T</i> can grow both under aerobic and anaerobic conditions. Furthermore, <i>G. auratiaca T-27T</i> genome encoded significantly large numbers of signal transduction components, sigma factors and transporters. These genomic features would provide an insight into the life style of <i>G. aurantiaca</i>, and facilitate isolation of not-yet-cultivated Gemmatimonadetes species. [bionite].<br> | |||
[[File:Genome_gemmatimonas.png|thumb|Genome structure of <i>G. aurantiaca</i> [Genome Information Broker]]] | |||
[[File:Genome_gemmatimonas.png|thumb|Genome structure of <i>G. aurantiaca</i> [Genome Information Broker]]] | |||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
<i>Gemmatimonas aurantiaca T-27T</i> possess a Gram-negative cell envelope, with the cytoplasmic and outer membranes readily visible. It is able to utilize substrates as sole carbon sources for energy. Some substrates are yeast extract, polypepton, acetate, and gelatin. When growth occurs catalase and oxidase are produced, while polyphosphate accumulation is present. [Genome Information Broker] | |||
==Ecology== | ==Ecology== | ||
<i>Gemmatimonas aurantiaca T-27T</i> is a very slow growing bacterium that needs specific factors to allow growth. Strain T-27T grows at 25–35 °C. No growth occurs at temperatures of below 20°C or above 37°C, while the optimum pH needed for growth is about pH 7, but it can grow with pH between 6.5 and 9.5. [Zhang et al.] | |||
==Pathology== | ==Pathology== | ||
Due to <i>G. aurantiaca</i> being so recently discovered, it is unknown at this time if this bacteria can cause harm to variety of hosts. Scientists hope to learn more about this bacteria and others like it in order to determine if they cause any harm to humans or any other host species. | |||
==Current Research== | ==Current Research== | ||
In 2003, Japanese scientists Hui Zhang, Yuji Sekiguchi, Satoshi Handa, Phillip Hugenholtz, Hongik Kim, Yoichi Kamagata, and Kazunori Nakamura were studying 16S rDNA sequences which had been previously studied by other scientists. They focused on a bacterium that was from enhanced biological phosphorus removal sludge. When isolating the T27T strain from the bacterium, they discovered slow growing circular, smooth, orange colonies. It took over 2 weeks of incubation to get any colonies at all, which could be the reason this bacterium has been over looked in the past. Many tests where performed, which allowed for many characteristics to be observed and recorded. It was determined by scientific standards that strain T27T is both novel and a distinctive bacterium. They hope this discovery will help identify more bacterium and will help to aide in the learning of many unknown pathways and/or production of products or enzymes. [Zhang et al.] | |||
In 2005, Davis et al. published a paper about bacteria growing in soil. Soils are inhabited by many bacteria from phylogentic groups. Most are poorly studied because it is very rare to isolate them in cultivation studies. The low frequencies with which bacterial cells from soil can form visible colonies, is due to them being unable to brow on microbiological media. In this study they used three different types of media and increased the incubation time to up to three months. This allowed the development of visible colonies and allowed for other scientists to begin researching unidentified species of bacteria. [Davis et al.] | |||
In 2009, <i>Gemmatimonas aurantiaca T27T</i> pigments were purified in a lab by Shinichi Takaichi, Takashi Maoka, Kazuto Takasaki, and Satoshi Hanada. They identified many carotenoids, some of which were not identified in other bacterium. They were able to identify the carotenoids and figure out the complete nucleotide sequences. With that they were able to propose a biosynthetic pathway for the carotenoids, as well as, the genes and enzymes needed for it. They also discovered that <i>G. aurantiaca</i> store these carotenoid pigments to function as antioxidants use as defense to high concentrations of oxygen. [Tamaki et al.] | |||
Also in 2009, cultures from soils revealed <i>G. aurantica</i> and Ellin5290. Both of which represent only one of at least four main lines of descent in the Gemmatimonadetes. This can be used to target genomic gaps in the bacterial and archaeal domains. [Hugenholtz et al.] | |||
==Cool Factor== | ==Cool Factor== | ||
What we found cool about <i>G. aurantiaca</i> is that its strain T27T is known to be novel. This is interesting because based on phenotypic tests that were done in the Zhang et al. study, it is known as physiologically unremarkable. The reason for this is because many diverse bacteria are gram negative aerobic heterotrophs, which this bacterium is not. Also, many bacteria can accumulate polyphosphate, which means this bacterium doesn't stand out in that aspect. But there are some ways this bacterium does stand out from other microbes which makes it novel. Its exotic ancestry comes from data collected by chemotaxonomic and ultrastructural tests. One is it has an unusual combination of fatty acids. The fatty acids aren't known to be in the type of bacterium found in this test. Also, the cell envelope is unusual because of the space between membranes is too wide for gram negative bacteria. Though much more is needed to determine just how novel strain T27T is, there is enough details that scientists can props this strain as being novel. [Zhang et al.] | |||
==References== | ==References== | ||
[http:// | 1. [http://gtps.ddbj.nig.ac.jp/single/index.php?spid=Gaur_T27 <i>Gemmatimonas aurantiaca T-27</i>." Genome Information Broker. Web.]<br> | ||
2. [http://ijs.sgmjournals.org/content/53/4/1155.full Zhang, Hui, Yuji Sekiguchi, Satoshi Hanada, Philip Hugenholtz, Hongik Kim, Yoichi Kamagata, and Kazunori Nakamura. "<i>Gemmatimonas Aurantiaca Gen. Nov.</i>, Sp. Nov., a Gram-negative, Aerobic, Polyphosphate-accumulating Micro-organism, the First Cultured Representative of the New Bacterial Phylum Gemmatimonadetes Phyl. Nov." International Journal of Systematic and Evolutionary Microbiology.]<br> | |||
3. http://www.bio.nite.go.jp/ngac/e/ga1-e.html<br> | |||
4. [http://mic.sgmjournals.org/content/156/3/757.full Shinichi Takaichi, Takashi Maoka, Kazuto Takasaki, and Satoshi Hanada "Carotenoids of <i>Gemmatimonas aurantiaca (Gemmatimonadetes): identification of a novel carotenoid, deoxyoscillol 2-rhamnoside, and proposed biosynthetic pathway of oscillol 2,2'-dirhamnoside</i>", Microbiology 2010 156: 757-763.]<br> | |||
5. [http://0-content.ebscohost.com.ilsprod.lib.neu.edu/pdf9/pdf/2009/EU9/01Mar09/36680400.pdf?T=P&P=AN&K=36680400&S=R&D=aph&EbscoContent=dGJyMMvl7ESeqLU4xNvgOLCmr0qep7VSsqy4S7SWxWXS&ContentCustomer=dGJyMPGrrk%2BvrLFOuePfgeyx43zx Hugenholtz, P., & Kyrpides, N. C. (2009). A changing of the guard. Environmental Microbiology, 11(3), 551-553. doi:10.1111/j.1462-2920.2009.01888.x]<br> | |||
6. [http://0-content.ebscohost.com.ilsprod.lib.neu.edu/pdf23_24/pdf/2009/EU9/01Jul09/42874819.pdf?T=P&P=AN&K=42874819&S=R&D=aph&EbscoContent=dGJyMMvl7ESeqLU4xNvgOLCmr0qep7VSsq%2B4Sa%2BWxWXS&ContentCustomer=dGJyMPGrrk%2BvrLFOuePfgeyx43zx Tamaki, H., Hanada, S., Sekiguchi, Y., Tanaka, Y., & Kamagata, Y. (2009). Effect of gelling agent on colony formation in solid cultivation of microbial community in lake sediment. Environmental Microbiology, 11(7), 1827-1834. doi:10.1111/j.1462-2920.2009.01907.x]<br> | |||
7. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC546801/?tool=pubmed Kathryn E. R. Davis, Shayne J. Joseph, Peter H. Janssen "Effects of Growth Medium, Inoculum Size, and Incubation Time on Culturability and Isolation of Soil Bacteria." Appl Environ Microbiol. 2005 February; 71(2): 826–834. doi: 10.1128/AEM.71.2.826-834.2005]<br> | |||
8. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC309996/?tool=pubmed Shayne J. Joseph, Philip Hugenholtz, Parveen Sangwan, Catherine A. Osborne, Peter H. Janssen "Laboratory Cultivation of Widespread and Previously Uncultured Soil Bacteria" Appl Environ Microbiol. 2003 December; 69(12): 7210–7215. doi: 10.1128/AEM.69.12.7210-7215.2003] | |||
Edited by student of Iris Keren: | Edited by student of Iris Keren: | ||
NEUS2012 Amal Mohamed, Soonji Kim, Jaclyn Egitto | NEUS2012 Amal Mohamed, Soonji Kim, Jaclyn Egitto | ||
Latest revision as of 00:14, 28 February 2012
A Microbial Biorealm page on the genus Gemmatimonas aurantiaca
Classification
Higher order taxa
Super Kingdom: Bacteria; Phylum: Gemmatimonadetes; Class: Gemmatimonadetes; Order: Gemmatimonadales; Family: Gemmatimonadaceae; Genus: Gemmatimonas
(NCBI)
Species
|
NCBI: Taxonomy |
Species: Gemmatimonas aurantiaca T-27T
Description and significance
The phylum Gemmatimonadetes is a very recently described bacterial group whose members are widespread in nature, particularly in soil habitats. There is only one cultured representative of this phylum that is Gemmatimonas aurantiaca. [Joseph et al.] G. aurantiaca T27T is an aerobic rod-shaped Gram-negative bacterium that is non-sporulating. It is motile by the use of flagella and it was isolated by anaerobic-aerobic sequential batch reactor under enhanced biological phosphorus removal conditions for wastewater treatment. G. aurantiaca T27T is a very slow growing bacterium and is a polyphosphate-accummulating bacteria. It is able to grow not only by anaerobic respiration under anaerobic conditions but also by oxygen respiration under aerobic conditions. It is also orange in color, whether growing on a plate or in a liquid media. Due to G. aurantiaca having many distinct phylogenetic traits, it suggests that this bacterium will have many unique enzymes and metabolic pathways. Because of its uniqueness it was promised that this bacterium would lead to the discovery of new carotenoids. [Takaichi et al.]
Genome structure
Gemmatimonas aurantiaca T-27T has a circular chromosome that consists of 4,636,964 base pairs of which majority, 64.28%, includes G/C pair. G. aurantiaca contains about 3,935 open reading frames, 48 tRNA genes and single rRNA operon were predicted. It also contains essential genes that were previously identified in model organisms such as Escherichia coli and Bacillus subtilis. This shows that their basic cellular system is not much different from other known microbes. G. aurantiaca T-27T can grow both under aerobic and anaerobic conditions. Furthermore, G. auratiaca T-27T genome encoded significantly large numbers of signal transduction components, sigma factors and transporters. These genomic features would provide an insight into the life style of G. aurantiaca, and facilitate isolation of not-yet-cultivated Gemmatimonadetes species. [bionite].
Cell structure and metabolism
Gemmatimonas aurantiaca T-27T possess a Gram-negative cell envelope, with the cytoplasmic and outer membranes readily visible. It is able to utilize substrates as sole carbon sources for energy. Some substrates are yeast extract, polypepton, acetate, and gelatin. When growth occurs catalase and oxidase are produced, while polyphosphate accumulation is present. [Genome Information Broker]
Ecology
Gemmatimonas aurantiaca T-27T is a very slow growing bacterium that needs specific factors to allow growth. Strain T-27T grows at 25–35 °C. No growth occurs at temperatures of below 20°C or above 37°C, while the optimum pH needed for growth is about pH 7, but it can grow with pH between 6.5 and 9.5. [Zhang et al.]
Pathology
Due to G. aurantiaca being so recently discovered, it is unknown at this time if this bacteria can cause harm to variety of hosts. Scientists hope to learn more about this bacteria and others like it in order to determine if they cause any harm to humans or any other host species.
Current Research
In 2003, Japanese scientists Hui Zhang, Yuji Sekiguchi, Satoshi Handa, Phillip Hugenholtz, Hongik Kim, Yoichi Kamagata, and Kazunori Nakamura were studying 16S rDNA sequences which had been previously studied by other scientists. They focused on a bacterium that was from enhanced biological phosphorus removal sludge. When isolating the T27T strain from the bacterium, they discovered slow growing circular, smooth, orange colonies. It took over 2 weeks of incubation to get any colonies at all, which could be the reason this bacterium has been over looked in the past. Many tests where performed, which allowed for many characteristics to be observed and recorded. It was determined by scientific standards that strain T27T is both novel and a distinctive bacterium. They hope this discovery will help identify more bacterium and will help to aide in the learning of many unknown pathways and/or production of products or enzymes. [Zhang et al.]
In 2005, Davis et al. published a paper about bacteria growing in soil. Soils are inhabited by many bacteria from phylogentic groups. Most are poorly studied because it is very rare to isolate them in cultivation studies. The low frequencies with which bacterial cells from soil can form visible colonies, is due to them being unable to brow on microbiological media. In this study they used three different types of media and increased the incubation time to up to three months. This allowed the development of visible colonies and allowed for other scientists to begin researching unidentified species of bacteria. [Davis et al.]
In 2009, Gemmatimonas aurantiaca T27T pigments were purified in a lab by Shinichi Takaichi, Takashi Maoka, Kazuto Takasaki, and Satoshi Hanada. They identified many carotenoids, some of which were not identified in other bacterium. They were able to identify the carotenoids and figure out the complete nucleotide sequences. With that they were able to propose a biosynthetic pathway for the carotenoids, as well as, the genes and enzymes needed for it. They also discovered that G. aurantiaca store these carotenoid pigments to function as antioxidants use as defense to high concentrations of oxygen. [Tamaki et al.]
Also in 2009, cultures from soils revealed G. aurantica and Ellin5290. Both of which represent only one of at least four main lines of descent in the Gemmatimonadetes. This can be used to target genomic gaps in the bacterial and archaeal domains. [Hugenholtz et al.]
Cool Factor
What we found cool about G. aurantiaca is that its strain T27T is known to be novel. This is interesting because based on phenotypic tests that were done in the Zhang et al. study, it is known as physiologically unremarkable. The reason for this is because many diverse bacteria are gram negative aerobic heterotrophs, which this bacterium is not. Also, many bacteria can accumulate polyphosphate, which means this bacterium doesn't stand out in that aspect. But there are some ways this bacterium does stand out from other microbes which makes it novel. Its exotic ancestry comes from data collected by chemotaxonomic and ultrastructural tests. One is it has an unusual combination of fatty acids. The fatty acids aren't known to be in the type of bacterium found in this test. Also, the cell envelope is unusual because of the space between membranes is too wide for gram negative bacteria. Though much more is needed to determine just how novel strain T27T is, there is enough details that scientists can props this strain as being novel. [Zhang et al.]
References
1. Gemmatimonas aurantiaca T-27." Genome Information Broker. Web.
2. Zhang, Hui, Yuji Sekiguchi, Satoshi Hanada, Philip Hugenholtz, Hongik Kim, Yoichi Kamagata, and Kazunori Nakamura. "Gemmatimonas Aurantiaca Gen. Nov., Sp. Nov., a Gram-negative, Aerobic, Polyphosphate-accumulating Micro-organism, the First Cultured Representative of the New Bacterial Phylum Gemmatimonadetes Phyl. Nov." International Journal of Systematic and Evolutionary Microbiology.
3. http://www.bio.nite.go.jp/ngac/e/ga1-e.html
4. Shinichi Takaichi, Takashi Maoka, Kazuto Takasaki, and Satoshi Hanada "Carotenoids of Gemmatimonas aurantiaca (Gemmatimonadetes): identification of a novel carotenoid, deoxyoscillol 2-rhamnoside, and proposed biosynthetic pathway of oscillol 2,2'-dirhamnoside", Microbiology 2010 156: 757-763.
5. Hugenholtz, P., & Kyrpides, N. C. (2009). A changing of the guard. Environmental Microbiology, 11(3), 551-553. doi:10.1111/j.1462-2920.2009.01888.x
6. Tamaki, H., Hanada, S., Sekiguchi, Y., Tanaka, Y., & Kamagata, Y. (2009). Effect of gelling agent on colony formation in solid cultivation of microbial community in lake sediment. Environmental Microbiology, 11(7), 1827-1834. doi:10.1111/j.1462-2920.2009.01907.x
7. Kathryn E. R. Davis, Shayne J. Joseph, Peter H. Janssen "Effects of Growth Medium, Inoculum Size, and Incubation Time on Culturability and Isolation of Soil Bacteria." Appl Environ Microbiol. 2005 February; 71(2): 826–834. doi: 10.1128/AEM.71.2.826-834.2005
8. Shayne J. Joseph, Philip Hugenholtz, Parveen Sangwan, Catherine A. Osborne, Peter H. Janssen "Laboratory Cultivation of Widespread and Previously Uncultured Soil Bacteria" Appl Environ Microbiol. 2003 December; 69(12): 7210–7215. doi: 10.1128/AEM.69.12.7210-7215.2003
Edited by student of Iris Keren: NEUS2012 Amal Mohamed, Soonji Kim, Jaclyn Egitto