Flavobacterium hibernum: Difference between revisions
No edit summary |
Slonczewski (talk | contribs) (→Author) |
||
| (74 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
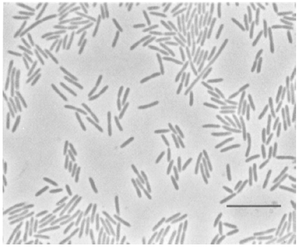
[[Image:Flav hib.png|thumb|300px|A photomicrograph of ''F. hibernum'' grown on nutrient-poor medium at 20C for 24hr (McCammon ''et al.'', 1998).]] | |||
==Classification== | ==Classification== | ||
'''Domain:''' ''Bacteria'' '''Phylum:''' ''Bacteroidetes'' '''Class:''' ''Flavobacteria'' '''Order:''' ''Flavobacteriales'' '''Family:''' ''Flavobacteriaceae'' | '''Domain:''' ''Bacteria'' | ||
'''Phylum:''' ''Bacteroidetes'' | |||
'''Class:''' ''Flavobacteria'' | |||
'''Order:''' ''Flavobacteriales'' | |||
'''Family:''' ''Flavobacteriaceae'' | |||
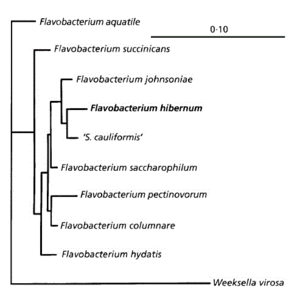
[[Image:FlavobacteriumhibernumPhylogeneticTree.gif|thumb|left|300px|This chart displays the evolutionary biology of ''F. hibernum'' (McCammon ''et al.'', 1998).]] | |||
===Species=== | ===Species=== | ||
| Line 8: | Line 37: | ||
{| | {| | ||
| height="10" bgcolor="#FFDF95" | | | height="10" bgcolor="#FFDF95" | | ||
'''NCBI: [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi? | '''NCBI: [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=37752 Taxonomy]''' | ||
|} | |} | ||
'' | '''''Flavobacterium hibernum''''' | ||
==Description and Significance== | ==Description and Significance== | ||
[[Image:Crooked Lake.jpg|thumb|Crooked Lake in Antarctica, the discovery site of ''F. hibernum''.]] | |||
''Flavobacterium hibernum'', meaning “wintry yellow bacteria”, is a Gram negative rod-shaped, aerobic bacterium, able to grow on cold, nutrient-poor media. First isolated from Crooked Lake in Antarctica (McCammon, 1998), but is now known to be found worldwide, in freshwater, ice, and soil ecosystems, and is largely unexplored genetically (Chen, 2006). An optimal growth temperature of 26ºC groups ''F. hibernum'' with psychrotrophic microbes, although the temperatures at which growth was possible ranged from -7ºC to 30ºC. This bacterium exhibits gliding motility, the mechanism of which is unknown. However, this method of motility may relate to its ability to lyse Gram-positive cells (such as ''E. coli'') as a bacteriovore in the food web. ''F. hibernum'' is able to utilize a wide variety of sugars and substrate containing other nutrients. | |||
Additionally, ''Flavobacterium hibernum'' and other ''Flavobacteria'' are commonly found in aquatic environments, including treeholes where container-breeding mosquitoes breed. ''Flavobacterium'' are consumed by several species of mosquito, including ''Anopheles gambiae'' and ''Aedes triseriatus'', which can act as vectors for disease (Chen, 2010). It has been proposed that ''F. hibernum'' or other members of its genus could be modified to deliver toxins from ''B. thuringiensis'' or ''B. sphaericus'' and be used as a “novel insecticide to lower mosquito populations” (Chen, 2010). | |||
==Genome Structure== | ==Genome Structure== | ||
Unlike other ''Flavobacterium'' species, the single circular chromosome of ''F. hibernum'' has not been fully sequenced, and is therefore not fully understood genetically. Only a few experiments have been documented displaying proteins utilized by this bacteria were noted, including the GyrB protein, which was used to identify diversity within the "Flavobacterium" family (Peeters and Willems, 2011), although little other conclusions were made. Other proteins include the SCH19 promoter, and many sections of the ribosomal RNA. Additionally, the GC content of the genome is 34 mol% (McCammon ''et al.,'' 1998). | |||
There has been a library of 9,000 clones containing chromosomal fragments of ''F. hibernum'' strain W22 in pSCH03 (promoter-trap plasmid). They go through as series of transcription and translation to ultimately prove that there are some promoter and ribosome binding site motifs of Flavobacterium strains (Chen ''et al.,'' 2007). | |||
==Cell Structure, Metabolism and Life Cycle== | ==Cell Structure, Metabolism and Life Cycle== | ||
''F. hibernum'' is a gram negative, rod-shaped, aerobic organism. Optimal temperature for growth of F. hibernum is 26˚C, but can grow from -7 to -31˚C, classifying it as a psychrotroph, and the typical size of the microbe is .7x1.8-13 µm. When grown at lower temperatures (~4˚C), a thick capsule forms. Both branched and hydroxyl fatty acids are found in ''F. hibernum''. This bacterium has potential use in dairy processing due to its β-galactosidase activity, which can hydrolyze lactose and make dairy products edible to those with lactose intolerance. The unique ability of this organism to do this at a low temperature keeps the dairy products from spoiling. Additionally, lactose can be used as a sole carbon source, and during lactose utilization ammonium ions are the nitrogen source. A variety of other sugars are also able to be utilized as carbon sources. Menaquinone 6, a variation of Vitamin K, is the chief respiratory molecule. Interestingly, this organism does not enter the microcyst lifestage as other organisms do to protect against environmental pressures, such as osmotic stress. However, as mentioned earlier, it is able to excrete extracellular polysaccharides to form gel-like matrix around a colony. This practice has been shown to reduce water loss. (McCammon ''et al.'') | |||
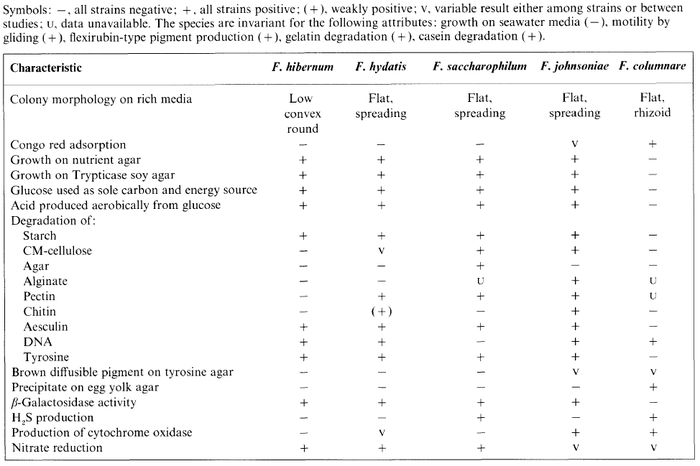
''F. hibernum'' is catalase positive, meaning it can decompose oxygen radicals, is unable to reduce oxygen using the enzyme oxidase, and is capable of reducing nitrate. It lacks the capability to produces hydrogen sulfide or indole and is unable to use citrate as a carbon source. The table below, from McCammon ''et al.'' (1998), gives detailed metabolic information about ''F. hibernum'' in comparison to other ''Flavobacterium'' species. | |||
[[Image:McCammon et al 1998 F. hibernum Metabolism Comparison.png|thumb|center|700px|This table compares metabolic processes of ''F. hibernum'' to other ''Flavobacterium sp.'' (McCammon ''et al.'', 1998)]] | |||
==Ecology and Pathogenesis== | ==Ecology and Pathogenesis== | ||
'''Ecology:''' | |||
''Flavobacterium hibernum'' has the ability to glide and lyse Gram-negative cells, which indicates that it could have the role of a bacterivore (McCammon, 1998). And since ''F. hibernum'' was originally discovered in a nutrient poor freshwater lake, being a bacterivore helps the ecosystem recycle resources from growing bacteria populations, establishing a balance in the amount of resources used (McCammon, 1998). It is also utilized as a food source by mosquito larvae, such as ''Aedes triseriatus'', a competent vector for La Crosse encephalitis virus (McJunkin and Minnich, 2009). | |||
Another focus of this microbe is the ability to reduce Nitrate to Nitrite; this can be helpful for creating a microbial loop. The microbial loop formed by ''F. hibernum'' is the primary route for the recycling of nutrients in these fresh water lakes found, not only in Antarctica, but all around the world (McCammon, 1998). | |||
''F. hibernum'' could also be isolated, at low temperatures ranging between 30 and 40 degrees Celsius, to trigger hydrolysis of lactose. This could possibly help aid in the processing of dairy foods because the human body cannot tolerate lactose found in unfermented dairy. ''F. hibernum'' is able to break down a large array of nutrients: casein, starch, aesculin, DNA, L-tyrosine, and Tween 80 (McCammon, 1998). | |||
'''Pathogenesis:''' | |||
Due to the fact that ''F. hibernum'' is a possible food source for mosquitoes in their larval stage, it is proposed that transferring of genes that encode for proteins toxic to mosquitoes, such as those from ''Bacillus thuringiensis'', to ''F. hibernum'' could be used as an effective insecticide (Chen, 2007). However, ''Flavobacterium'' are so diverse that the conventional transfer of genes, by proteobacteria plasmids, selectable markers, or transposons, and thus are difficult manipulate genetically (Chen, 2007). Experiments are currently being performed to determine the optimal way to transfer genes to ''Flavobacterium''. A potential drawback to this approach stems from the toxicity of certain strains of ''Flavobacterium'' to freshwater fish (Bernardet ''et al.,'', 1996) | |||
==References== | ==References== | ||
[http://ijs.sgmjournals.org/content/46/1/128.full.pdf J.F. Bernardet, Segers, P., Vancanneyt, M., Berthe, F., Kersters, K., and Vandamme, P. "Cutting a Gordian Knot: Emended Classification and Description of the Genus Flavobacterium, Emended Description of the Family Flavobacteriaceae, and Proposal of ''Flavobacterium hydatis'' norn. nov." ''International Journal of Systematic Bacteriology'', 1996, p. 128-148.] | |||
[http://ac.els-cdn.com/S0378111910000648/1-s2.0-S0378111910000648-main.pdf?_tid=c3fe9ec7e37e56b9418dad741a782276&acdnat=1334540997_519b6b2f011a8aa9b578bd532a735d0c S. Chen, Kaufman M.G., Bagdasarian M., Bates A.K., Walker ED. "Development of an Efficient Expression System for Flavobacterium Strains." ''Gene'', 458, 2010: 1-10. ASFA: Aquatic Sciences and Fisheries Abstracts.] | |||
[http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2006.00543.x/pdf S. Chen, Bagdasarian, M., Kaufman, M.G., and Walker, E.D. "Organization of a partial S10 operon and its transcriptional analysis in ''Flavobacterium hibernum'' strain W22". ''FEMS Microbiology Letters'', 2007, Volume 267, Issue 1, pages 38–45.] | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1828668/?tool=pubmed S. Chen, Bagdasarian, M., Kaufman, M.G., and Walker, E.D. "Characterization of strong promoters from an environmental Flavobacterium hibernum strain by using a green fluorescent protein-based reporter system." ''Applied and Environmental Microbiology'', 2007. 73(4): 1089–1100.] | |||
[http://ijs.sgmjournals.org/content/48/4/1405.full.pdf S.A. McCammon, B.H. Innes, J.P. Bowman, P.D. Franzmann, S.J. Dobson, P.E. Holloway, J.H. Skerratt, P.D. Nichols, and L.M. Rankin. "''Flavobacterium hibernum'' sp. nov., a lactose-utilizing bacterium from a freshwater Antarctic lake". ''International Journal of Systematic and Bacteriology''. 1998. Volume 48. p. 1405-1412.] | |||
McJunkin JE, Minnich LL. La Crosse encephalitis and other California serogroup viruses. In: Feign RD, Cherry JD, eds. Textbook of pediatric infectious diseases. 6th ed. Philadelphia, PA: Saunders; 2009:2557--68. | |||
[http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2011.02326.x/abstract K. Peeters and A. Willems. "The gyrB gene is a useful phylogenetic marker for exploring the diversity of Flavobacterium strains isolated from terrestrial and aquatic habitats in Antarctica". ''FEMS Microbiology Letters'', 2011. | |||
Volume 321, Issue 2, pages 130–140.] | |||
[http://www.mrl.nott.ac.uk/~mbf/antarctica/photo/crooked.jpg Photograph of Crooked Lake, taken from the University of Nottingham's Mixed Reality Lab.] | |||
[http://myrmecos.net/insects/AedesTriseriatus9.html Photograph of ''Aedes triseriatus'', taken from Alex Wild.] | |||
==Author== | ==Author== | ||
Page authored by | Page authored by Geoffrey Grzesiak, Matthew Herman, Andrea Hoffman, and Megan Hofmeister, students of [http://www.kbs.msu.edu/faculty/lennon/ Prof. Jay Lennon] at Michigan State University. | ||
[[Category:Pages edited by students of Jay Lennon at Michigan State University]] | |||
Latest revision as of 13:40, 4 June 2014
Classification
Domain: Bacteria
Phylum: Bacteroidetes
Class: Flavobacteria
Order: Flavobacteriales
Family: Flavobacteriaceae
Species
|
NCBI: Taxonomy |
Flavobacterium hibernum
Description and Significance
Flavobacterium hibernum, meaning “wintry yellow bacteria”, is a Gram negative rod-shaped, aerobic bacterium, able to grow on cold, nutrient-poor media. First isolated from Crooked Lake in Antarctica (McCammon, 1998), but is now known to be found worldwide, in freshwater, ice, and soil ecosystems, and is largely unexplored genetically (Chen, 2006). An optimal growth temperature of 26ºC groups F. hibernum with psychrotrophic microbes, although the temperatures at which growth was possible ranged from -7ºC to 30ºC. This bacterium exhibits gliding motility, the mechanism of which is unknown. However, this method of motility may relate to its ability to lyse Gram-positive cells (such as E. coli) as a bacteriovore in the food web. F. hibernum is able to utilize a wide variety of sugars and substrate containing other nutrients. Additionally, Flavobacterium hibernum and other Flavobacteria are commonly found in aquatic environments, including treeholes where container-breeding mosquitoes breed. Flavobacterium are consumed by several species of mosquito, including Anopheles gambiae and Aedes triseriatus, which can act as vectors for disease (Chen, 2010). It has been proposed that F. hibernum or other members of its genus could be modified to deliver toxins from B. thuringiensis or B. sphaericus and be used as a “novel insecticide to lower mosquito populations” (Chen, 2010).
Genome Structure
Unlike other Flavobacterium species, the single circular chromosome of F. hibernum has not been fully sequenced, and is therefore not fully understood genetically. Only a few experiments have been documented displaying proteins utilized by this bacteria were noted, including the GyrB protein, which was used to identify diversity within the "Flavobacterium" family (Peeters and Willems, 2011), although little other conclusions were made. Other proteins include the SCH19 promoter, and many sections of the ribosomal RNA. Additionally, the GC content of the genome is 34 mol% (McCammon et al., 1998).
There has been a library of 9,000 clones containing chromosomal fragments of F. hibernum strain W22 in pSCH03 (promoter-trap plasmid). They go through as series of transcription and translation to ultimately prove that there are some promoter and ribosome binding site motifs of Flavobacterium strains (Chen et al., 2007).
Cell Structure, Metabolism and Life Cycle
F. hibernum is a gram negative, rod-shaped, aerobic organism. Optimal temperature for growth of F. hibernum is 26˚C, but can grow from -7 to -31˚C, classifying it as a psychrotroph, and the typical size of the microbe is .7x1.8-13 µm. When grown at lower temperatures (~4˚C), a thick capsule forms. Both branched and hydroxyl fatty acids are found in F. hibernum. This bacterium has potential use in dairy processing due to its β-galactosidase activity, which can hydrolyze lactose and make dairy products edible to those with lactose intolerance. The unique ability of this organism to do this at a low temperature keeps the dairy products from spoiling. Additionally, lactose can be used as a sole carbon source, and during lactose utilization ammonium ions are the nitrogen source. A variety of other sugars are also able to be utilized as carbon sources. Menaquinone 6, a variation of Vitamin K, is the chief respiratory molecule. Interestingly, this organism does not enter the microcyst lifestage as other organisms do to protect against environmental pressures, such as osmotic stress. However, as mentioned earlier, it is able to excrete extracellular polysaccharides to form gel-like matrix around a colony. This practice has been shown to reduce water loss. (McCammon et al.)
F. hibernum is catalase positive, meaning it can decompose oxygen radicals, is unable to reduce oxygen using the enzyme oxidase, and is capable of reducing nitrate. It lacks the capability to produces hydrogen sulfide or indole and is unable to use citrate as a carbon source. The table below, from McCammon et al. (1998), gives detailed metabolic information about F. hibernum in comparison to other Flavobacterium species.
Ecology and Pathogenesis
Ecology: Flavobacterium hibernum has the ability to glide and lyse Gram-negative cells, which indicates that it could have the role of a bacterivore (McCammon, 1998). And since F. hibernum was originally discovered in a nutrient poor freshwater lake, being a bacterivore helps the ecosystem recycle resources from growing bacteria populations, establishing a balance in the amount of resources used (McCammon, 1998). It is also utilized as a food source by mosquito larvae, such as Aedes triseriatus, a competent vector for La Crosse encephalitis virus (McJunkin and Minnich, 2009).
Another focus of this microbe is the ability to reduce Nitrate to Nitrite; this can be helpful for creating a microbial loop. The microbial loop formed by F. hibernum is the primary route for the recycling of nutrients in these fresh water lakes found, not only in Antarctica, but all around the world (McCammon, 1998).
F. hibernum could also be isolated, at low temperatures ranging between 30 and 40 degrees Celsius, to trigger hydrolysis of lactose. This could possibly help aid in the processing of dairy foods because the human body cannot tolerate lactose found in unfermented dairy. F. hibernum is able to break down a large array of nutrients: casein, starch, aesculin, DNA, L-tyrosine, and Tween 80 (McCammon, 1998).
Pathogenesis: Due to the fact that F. hibernum is a possible food source for mosquitoes in their larval stage, it is proposed that transferring of genes that encode for proteins toxic to mosquitoes, such as those from Bacillus thuringiensis, to F. hibernum could be used as an effective insecticide (Chen, 2007). However, Flavobacterium are so diverse that the conventional transfer of genes, by proteobacteria plasmids, selectable markers, or transposons, and thus are difficult manipulate genetically (Chen, 2007). Experiments are currently being performed to determine the optimal way to transfer genes to Flavobacterium. A potential drawback to this approach stems from the toxicity of certain strains of Flavobacterium to freshwater fish (Bernardet et al.,, 1996)
References
McJunkin JE, Minnich LL. La Crosse encephalitis and other California serogroup viruses. In: Feign RD, Cherry JD, eds. Textbook of pediatric infectious diseases. 6th ed. Philadelphia, PA: Saunders; 2009:2557--68.
[http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2011.02326.x/abstract K. Peeters and A. Willems. "The gyrB gene is a useful phylogenetic marker for exploring the diversity of Flavobacterium strains isolated from terrestrial and aquatic habitats in Antarctica". FEMS Microbiology Letters, 2011. Volume 321, Issue 2, pages 130–140.]
Photograph of Crooked Lake, taken from the University of Nottingham's Mixed Reality Lab.
Photograph of Aedes triseriatus, taken from Alex Wild.
Author
Page authored by Geoffrey Grzesiak, Matthew Herman, Andrea Hoffman, and Megan Hofmeister, students of Prof. Jay Lennon at Michigan State University.