Bacillus anthracis as a Bioterrorism Agent: Difference between revisions
| (110 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | {{Uncurated}} | ||
==Introduction== | ==Introduction== | ||
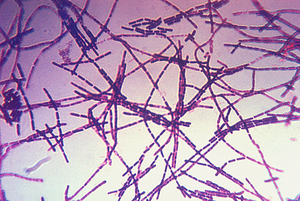
[[Image:Bacillus_anthracis.png|thumb|300px|right|Figure 1. Photomicrograph of a Gram-stain of <i>B. anthracis</i> which is the causative agent of anthrax disease. [http://commons.wikimedia.org/wiki/File:Bacillus_anthracis_Gram.jpg Image from the CDC Public Health Library.]]] | |||
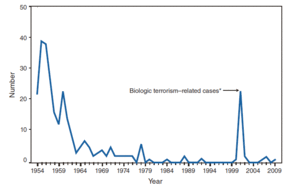
<i>Bacillus anthracis</i> is the causative agent of anthrax and has been adapted for use in bioterrorism. It is a large Gram-positive aerobic, rod shaped, bacillus bacterium (Figure 1). It ranges in size from 1-1.5 x 3-10 µm and is the only obligate pathogen within the genus <i>bacillus</i>.[http://www.ncbi.nlm.nih.gov/pubmed/11173038<sup>1</sup>] Anthrax is an acute disease, that appears suddenly and progresses rapidly, affecting humans and animals. Previously anthrax was perceived as a damaging disease affecting domesticated animals, the farming economy, and some industrial workers; however there has been a shift to now view the disease as an agent of bioterrorism. On October 5, 2001 the first anthrax related death in the United States occurred in over 25 years in which <i>B. anthracis</i> spores were mailed. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1769905/<sup>2</sup>] <i>B. anthracis</i> took the form of assymetric warfare where tactics, resources, and potential for destruction was unknown. According to the Center for Disease Control, approximately 1-2 cases of Anthrax occur each year; however in 2001, 22 cases of anthrax total occurred in the United States due to this act of bioterrorism (Figure 2).[http://www.bt.cdc.gov/agent/anthrax/faq/<sup>3</sup>] | |||
[[Image:Anthrax_cases_by_year.gif|thumb|300px|right|Figure 2. Number of Anthrax cases in the United States from 1954-2009. Highlighted are the bioterrorism related incidents of anthrax, which occurred 2001-2002. [http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5853a1.htm Image from the CDC Public Health Library.]]] | |||
<br> | <br><i>B. anthracis</i> is successful as a bioterrorism agent due to its resistance to harsh environmental conditions through its ability to exist in a spore state. With the potential to have extreme and damaging effects on the population, the study of <i>B. anthracis</i> and the discovery of rapid and reliable technologies to identify the species is of utmost concern. | ||
== | ==Biology and Genetics of <i> B. anthracis</i>== | ||
[[ | ====Virulence Plasmids==== | ||
Two virulence plasmids, pXO1 and pXO2, encode the major virulence factors for <i>B. anthracis</i>.[http://www.ncbi.nlm.nih.gov/pubmed/10594977<sup>4</sup>] Plasmid pXO1 codes for three toxins which cause [http://en.wikipedia.org/wiki/Bleeding hemorrhage], [http://en.wikipedia.org/wiki/Edema edema], and [http://en.wikipedia.org/wiki/Necrosis necrosis] and is 184.5 kilobase pairs (kbp) in size. The exotoxins it codes for are binary, which include the lethal factor (LF) and edema factor (EF), and the protein known as the protective antigen (PA). Therefore, the protective antigen it codes for acts as a binding domain on the host to permit entry of the toxin. pXO2 is a capsule bearing plasmid that is smaller than pXO1 having 95.3 kbp in size. It encodes three genes (<i>cap A, cap B</i>, and <i>cap C</i>) which are involved in the synthesis of the polyglutamyl capsule. The capsule prevents the bacteria from phagocytosis and is poorly recognized by the immune system of the host.[http://www.nejm.org/doi/full/10.1056/NEJM199909093411107<sup>5</sup>] Both pXO1 and pXO2 plasmids are necessary to provide the anthrax toxin and capsule respectively for <i>B. anthracis</i> to have full virulence. An attenuated strain results when either plasmid is lost.[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1769905/<sup>6</sup>] | |||
< | ====Three Proteins of Anthrax==== | ||
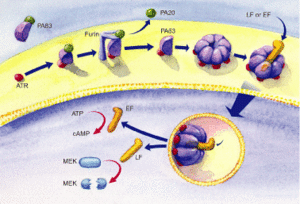
[[Image:Mechanism of Anthrax.gif|thumb|300px|right|Figure 3. <i>B. anthracis</i> secretes three proteins to form the Anthrax toxin. PA83 binds to the Anthrax Toxin Receptor on the cell surface and is cleaved by Furin to produce PA63. LF and EF internalize once PA63 forms a heptameric structure to spread the disease throughout the host. [http://www.rndsystems.com/cb_detail_objectname_FA03_Bacillus.aspx Image from R&D systems.]]] | |||
The protective antigen (PA), lethal factor (LF), and edema factor (EF) are proteins that comprise the anthrax toxin. They are encoded by the genes <i>pag, lef,</i> and <i>cya</i>, respectively. The PA factor contains the cell-binding domain. LF or EF are responsible for cell toxicity by gaining access to the host cell's cytoplasms. This is able to occur when PA locates specific host cell surface receptors and cleavage by [http://en.wikipedia.org/wiki/Furin Furin], a cell surface protein, ensues. After cleavage, the remaining part of the PA auto-assembles in the cell membrane to form a heptameric (seven-membered) pore. The other protein components, EF or LF, then bind to the PA. PA enables the EF or LF toxins to then be released into the host cell's cytoplasm to cause infection (Figure 3). Once EF enters the cytoplasm, it raises [http://en.wikipedia.org/wiki/Cyclic_adenosine_monophosphate cyclic adenosine monophosphate (cAMP)] levels. This causes an inhibition of immune cell function, specifically resulting in edema, which is commonly referred to as swelling in which there is an accumulation of fluid trapped in a particular part of the body's tissues. If LF is released into the hosts' cells, it cleaves the hosts' [https://en.wikipedia.org/wiki/Protein_kinase protein kinases], which leads to altered signaling pathways and ultimately results in cell death. [http://www.ncbi.nlm.nih.gov/pubmed/15680766/<sup>7</sup>] | |||
====Spore and Vegetative State==== | |||
<i>B. anthracis </i> is a bacterium that can exist in both a spore and vegetative state, which make it a favorable agent for bioterrorism. A spore is a dormant and resilient structure that can exist for extended periods of time with a lack of nutrients. <i>B. anthracis</i> can form spores that are resilient to harsh environments, such as ultraviolet and ionizing radiation, heat, and various chemicals.[http://onlinelibrary.wiley.com/store/10.1111/j.1574-6941.1997.tb00399.x/asset/j.1574-6941.1997.tb00399.x.pdf?v=1&t=her83r5w&s=18cf9c1b9e1475edacccd98768c494abb4dd8219<sup>8</sup>] A spore can germinate to exist in its vegetative state when the environmental conditions are favorable. To become a spore again, sporulation occurs, thereby ensuring the survival of <i>B. anthracis</i> during a destructive environmental condition. | |||
==Routes of Infection== | |||
< | Anthrax has three forms in which virulence occurs, which include cutaneous anthrax, gastrointestinal anthrax, and inhalational anthrax. When the host is exposed to the spores of <i>B. anthracis</i> via one of these three routes, infection occurs. | ||
====Cutaneous Anthrax==== | |||
Cutaneous anthrax comprises over 90% of all human cases. It is obtained via a lesion on the skin in which infection occurs through an abrasion, cut, or insect bite.[http://www.ncbi.nlm.nih.gov/pubmed/10594977<sup>9</sup>] This type of anthrax manifests itself through a black [http://en.wikipedia.org/wiki/Eschar eschar], which is dead tissue present on the surface of the skin as a scab, and is associated with [http://en.wikipedia.org/wiki/Edema edema]. Some patients also experience symptoms of swollen lymph nodes, fever, and headache. | |||
Anthrax | ====Gastrointestinal Anthrax==== | ||
Gastrointestinal anthrax results from the ingestion of undercooked meat from animals who have <i>B. anthracis</i> in its vegetative form or dormant spore state. This form of the disease has a high mortality rate because it is difficult to make an early diagnosis due to its’ non specific presentation. Symptoms may include nausea, vomiting, appetite loss, fever, sore throat, and a swollen neck. | |||
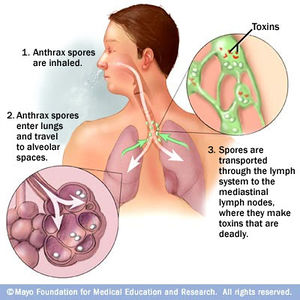
[[Image:Inhalation_Anthrax_Pathway.jpg|thumb|300px|right|Figure 4. The pathway in which anthrax spores spread to the human body following inhalation. [http://www.mayoclinic.com/health/medical/IM03225 Image from the Mayo Clinic].]] | |||
== | ====Inhalation Anthrax==== | ||
Inhalation anthrax is another form in which anthrax can be contracted. This route of infection is particularly relevant for the contraction of anthrax when used as an agent of bioterrorism. Traditionally, inhalation anthrax was associated with industrial exposure as spores in the textile, meat packing, leather-tanning, bone meal processing, and hair/wool-sorting industries; however it became associated with bioterrorism in 2001 when anthrax was included in envelopes mailed within the United States. Beginning with “flu-like” symptoms of mild fever, fatigue, [http://en.wikipedia.org/wiki/Malaise malaise] (discomfort associated with illness or infection), [http://en.wikipedia.org/wiki/Myalgia myalgia] (muscle pain), and non-productive cough from two to five days after initial exposure, the disease develops into an acute illness characterized by [http://en.wikipedia.org/wiki/Dyspnea dyspnea] (shortness of breath), [http://en.wikipedia.org/wiki/Stridor stridor] (wheezing caused by a narrowed or obstructed airway), fever, [http://en.wikipedia.org/wiki/Cyanosis cyanosis] (bluish discoloration of skin), and [http://en.wikipedia.org/wiki/Pleural_effusion pleural effusion] (excess fluid surrounding the lungs). With the initial display of "flu-like" symptoms when <i>B. anthracis</i> is inhaled, it difficult to diagnose the disease as anthrax. Anthrax manifests itself in the human body after inhalation of spores by first traveling to alveolar spaces within the lungs. Then the spores are transported through the [https://en.wikipedia.org/wiki/Lymphatic_system lymphatic system] to infect the entire body (Figure 4). Once the infection is established, it becomes refractory to treatment; therefore, the success of medical intervention is minimal. However, if the infection is diagnosed very early on, the disease may be successfully treated by multidrug antibiotic regimens and supported care; yet the prognosis of patients with inhalational anthrax is dismal as the mortality rate is high.[http://www.ncbi.nlm.nih.gov/pubmed/10594977<sup>10</sup>] | |||
==Methods of Detection== | |||
[[Image:Methods_Table.png|thumb|300px|right|Figure 5. Summary of selected detection methods to identify <i>B. anthracis</i>. Adapted from [http://www.ncbi.nlm.nih.gov/pubmed/20399814 Rao, S. et al, 2010] and [http://www.ncbi.nlm.nih.gov/pubmed/16283259 Edwards, K., and Clancy, H., 2005]]] | |||
A variety of methods exist for identifying <i>B. anthracis</i>. Phenotypic, antibody, and nucleic acid based methods are current techniques to identify the toxin in its various conditions including as a spore or in a vegetative state as well as using the microorganisms' DNA (Figure 5). | |||
====Conventional Phenotypic Methods of Identification==== | |||
<br><i>B. anthracis</i> can be detected using conventional microbiological methods via phenotypic identification. The organism selectively grows on polymyxin-lysozyme EDTA-thallous acetate (PELT) agar.[http://www.ncbi.nlm.nih.gov/pubmed/10475956<sup>11</sup>] However, this method requires a 1-2 day incubation period and requires additional tests to confirm the presence of the disease. Additionally, <i>B. anthracis</i> can be visualized using McFadyean’s capsule-staining test which uses McFadyean’s polychrome methylene blue to stain the capsule and rods.[http://www.nature.com/nature/journal/v393/n6687/full/393747a0.html<sup>12</sup>] Yet, these traditional methods for phenotypic identification of <i>B. anthracis</i> tend to be the most laborious and frequently require 1-2 days of testing. Laboratory personnel require additional training and certification to accurately identify this uncommon organism, which makes these methods impractical. Additionally, to confirm the presence of the pathogen, suspect isolates must frequently be further characterized through molecular techniques.[http://www.ncbi.nlm.nih.gov/pubmed/20399814<sup>13</sup>] | |||
====Antibody Based Methods of Identification==== | |||
<br>Immunoradiometric assays, IRMA, utilizes an immunofluorescence system which acts to detect antigens displayed on the surface of a spore. This method can immobilize spores in order measure concentrations of antigens through the amount of antibody used. Typically, immunofluorescence assays provide a rapid detection of spores; however, specific [http://en.wikipedia.org/wiki/Serology serological] detection is complex as a result of cross-reactivity between polyclonal and monoclonal antibodies and antigens as well as between <i>B. anthracis</i> and closely related species such as <i>B. cereus</i>. Despite these complications, the technique of immunoradiometric assays has been proven to successfully identify unique <i>B. anthracis</i> spore surface antigens. This was determined as a result of the lack of cross-reactivity with <i>B. cereus</i>, a closely related species.[http://www.ncbi.nlm.nih.gov/pubmed/6418708<sup>14</sup>] However, because the antibody must be radiolabeled, this method has been supplanted with the enzyme-linked immunosorbent assay. | |||
<br>< | <br>The [http://en.wikipedia.org/wiki/ELISA enzyme-linked immunosorbent assay], ELISA, detects immobilized spores of <i>B. anthracis</i> through identifying the spore surface antigens. Similar to an IRMA, an ELISA quantifies the amount of antibodies or proteins present in a sample; however unlike the IRMA, the ELISA method is a non-radioactive technique. The antigen must be immobilized when the ELISA is performed on a 96-well plate to permit high throughput results. For a successful ELISA, there must be a highly specific antibody-antigen interaction, which has been accomplished in the analysis of <i>B. anthracis</i> species. Antibodies displayed no cross-reactivity to other members of the <i>B. cereus</i> group, which is closely related to <i>B. anthracis</i>. Therefore, this technique is a reliable and effective technique for the identification of the <i>B. anthracis</i> species.[http://www.ncbi.nlm.nih.gov/pubmed/19793896<sup>15</sup>] | ||
<br><i>B. anthracis</i> | <br>[http://en.wikipedia.org/wiki/Flow_cytometry Flow cytometry] has also been used to detect <i>B. anthracis</i> spores through fluorescently labeled antibodies. A single wavelength of light is used to excite the fluorescent labels to disntiguish the entity of interest. The technique suspends cells in a stream of fluid and directs them through an electronic detection apparaturs. Flow cytometry is advantangeous because it allow the concurrent detection of multiple characteristics of particles. Quantum dots, which have narrower emission peaks than typical fluorophores used in flow cytometry, can be used in the detection <i>B. anthracis</i>. They result in an increase in accuracy of detecting cells in real-time and over extended periods of time. The quantum dots cause an increase in brightness and report more consistent results than traditional fluorophores. Although flow cytometry is a rapid method, it has a low limit of detection of 10<sup>3</sup> spores, therefore samples must be in abundance, which is not ideal.[http://www.ncbi.nlm.nih.gov/pubmed/11084608 <sup>16</sup>] | ||
<br>< | ====Nucleic Acid Based Methods of Identification==== | ||
<br>Nucleic acid based methods, including PCR, Real-Time PCR, and multiplex PCR, for the detection of <i>B. anthracis</i> rely on the selection of nucleic acid probes that are specific to the species and must have no cross-reactivity with other species. Primers frequently target amplification of the genes, <i>lef, pag,</i> and <i>cap</i> located on either or both of the virulence plasmids pXO1 and pXO2. [http://www.ncbi.nlm.nih.gov/pubmed/16283259<sup>17</sup>] | |||
<br> | <br>Using [http://en.wikipedia.org/wiki/Pcr PCR], strains containing the plasmids pXO1 and pXO2 are amplified. Analysis by gel electrophoresis follows as well as DNA sequencing. These methods can provide a measure of the genetic variability between isolates. The sequence <i>rpoB</i> is commonly used in PCR amplification of <i>B. anthracis</i>. The 175 base-pair sequence is highly selective for <i>B. anthracis</i> because it has the ability to discriminate <i>B. anthracis</i> from other <i>Bacillus</i> strains. Only one strain out of 175 tested bacilli strains has been found to be cross-reactive.[http://aem.asm.org/content/67/8/3720<sup>18</sup>] Ultimately, PCR is able to detect the presence of both virulence plasmids, can characterize <i>B. anthracis</i> vaccine strains, and detect spores from soil samples. | ||
<br> | <br>[http://en.wikipedia.org/wiki/QPCR Real-Time PCR], also known as quantitative PCR (qPCR), utilizes fluorescence to detect the DNA it amplifies. Fluorescence increases in a real-time manner as additional copies of DNA are produced. This type of assay avoids the need for further processing of the DNA to detect and quantify amplified sequences separately as necessary with PCR. Additionally, amplification is observed in real-time rather than after the fact. Since amplification and detection occur in the same chamber, there is a reduced likelihood for cross-contamination of products. In the 2001 anthrax outbreak in the United States in which <i>B. anthracis</i> was used as an agent of bioterrorism, real-time PCR was used to test and compare 542 isolates from clinical, environmental, and suspect powder samples from both this outbreak and prior outbreaks.[http://wwwnc.cdc.gov/eid/article/8/10/02-0393_article.htm<sup>19</sup>] Every hour, five copies per real-time PCR reaction of <i>B. anthracis</i> was detected and amplified. Before amplification occurred, the method required a growth period for 18-24 hours and an extraction time of 0.5-1 hour.[http://www.ncbi.nlm.nih.gov/pubmed/12149348<sup>20</sup>] 100% specificity was reported using this type of assay even when 56 non <i>B. anthracis</i> species were tested. | ||
<br> | <br>[http://en.wikipedia.org/wiki/Multiplex_polymerase_chain_reaction Multiplex PCR] amplifies specific regions of DNA using numerous sets of primers in a single reaction tube. By amplifying and detecting multiple target sequences on DNA, multiplex PCR is beneficial in eliminating false-negative and false-positive results.[http://www.ncbi.nlm.nih.gov/pubmed/15522596<sup>21</sup>] | ||
[ | <br>These amplification methods are often combined with high-throughput techniques for sample preparation and detection. Nucleic acid based systems are more sensitive than antigen and antibody-based detection systems in that they have the ability to detect fewer microorganisms in shorter amounts of time.[http://www.ncbi.nlm.nih.gov/pubmed/12588551<sup>22</sup>] Each method of PCR amplification has their own unique benefits for amplifying DNA; however, not all techniques are ideal to detect low levels of bacteria in environmental samples. For example, the downfall of real-time PCR is that this technique requires a DNAse step to remove DNA to prevent amplification of both DNA and RNA. The DNAse step to remove DNA has been proven to not be completely effective; therefore additional complex and time-consuming steps are necessary to ensure accurate results. | ||
< | ==Conclusion== | ||
<i>B. anthracis</i> has many genetic similarities among various <i>Bacillus</i> species and can exist in the spore and vegetative state making it an ideal agent for bioterrorism. These features make the detection and specific identification of <i>B. anthracis</i> difficult and have proven to require complex techniques and laborious methods. However, targeting the virulence plasmids pXO1 and pXO2 of <i>B. anthracis</i> have successfully been used in detecting the species. As an agent for a potential biological threat, <i>B. anthracis</i> is more frequently being studied. Due to the heightened interest in the organism, there have been a large number of detection technologies that have recently been developed and are underdevelopment to reliably and quickly characterize and identify the species with the hopes of controlling anthrax outbreaks and treating the outbreaks rapidly. | |||
==References== | |||
< | <sup>1</sup>[http://www.ncbi.nlm.nih.gov/pubmed/11173038 Baillie, L., Read, T.D., "<i>Bacillus anthracis</i>: a bug with attitude". <i>Current Opinion Microbiology</i>. 2001. Volume 4. p 78-81.] | ||
< | <sup>2</sup>[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1769905/ Spencer, R.C. "<i>Bacillus anthracis</i>". <i>Journal of Clinical Pathology</i>. 2003. Volume 56. p. 182-187.] | ||
< | <sup>3</sup>[http://www.bt.cdc.gov/agent/anthrax/faq/ "Questions and answers about Anthrax". <i>Center for Disease Control and Prevention.</i>. 2012.] | ||
< | <sup>4</sup>[http://www.ncbi.nlm.nih.gov/pubmed/10594977 Little, S.F. "Molecular pathogenesis of <i>Bacillus anthracis</i> infection". <i>Microbes and Infection</i>. 1999. Volume 1. 131-139.] | ||
== | <sup>5</sup>[http://www.nejm.org/doi/full/10.1056/NEJM199909093411107 Dixon, T., Meselson, M., Guillemin, J., and Hannah, P., "Anthrax". <i>New England Journal of Medicine</i> 1999. Volume 341. p. 815-826.] | ||
<sup>6</sup>[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1769905/ Spencer, R.C. "<i>Bacillus anthracis</i>". <i>Journal of Clinical Pathology</i>. 2003. Volume 56. 182-187.] | |||
<sup>7</sup>[http://www.ncbi.nlm.nih.gov/pubmed/15680766 Abramj, L., Reign, N., and van der Gopt, F., "Anthrax Toxin: The Long and Winding Road that Leads to the Kill", <i>Trends in Microbiology</i> 2005. Volume 13. p. 72-78] | |||
<sup>8</sup>[http://onlinelibrary.wiley.com/store/10.1111/j.1574-6941.1997.tb00399.x/asset/j.1574-6941.1997.tb00399.x.pdf?v=1&t=her83r5w&s=18cf9c1b9e1475edacccd98768c494abb4dd8219 Sjostedt, A., Eriksson, U., Ramisse, V., and Garrigue, H. "Detection of <i>Bacillus anthracis</i> spores in soil by PCR". <i>Microbial Ecology</i>. 1997. Volume 23. p. 159-168.] | |||
< | <sup>9</sup>[http://www.ncbi.nlm.nih.gov/pubmed/10594977 Little, S.F. "Molecular pathogenesis of <i>Bacillus anthracis</i> infection". <i>Microbes and Infection</i>. 1999. Volume 1: 131-139.] | ||
<sup>10</sup> [http://www.ncbi.nlm.nih.gov/pubmed/10594977 Little, S.F. "Molecular pathogenesis of <i>Bacillus anthracis</i> infection" <i>Microbes and Infection</i>. 1999. Volume 1. p. 131-139.] | |||
<sup>11</sup>[http://www.ncbi.nlm.nih.gov/pubmed/10475956 Turnbull P. "Definitive identification of <i>Bacillus anthracis</i>--a review" <i>Journal of Applied Microbiology</i> 1999. Volume 87. p. 237-240.] | |||
<sup>12</sup>[http://www.nature.com/nature/journal/v393/n6687/full/393747a0.html Redmond, C., Pearce, M., Manchee, R., and Berdal, B. "Deadly relic of the great war". <i>Nature</i> 1998. Volume 393. p. 747-748.] | |||
<sup> | <sup>13</sup>[http://www.ncbi.nlm.nih.gov/pubmed/20399814 Rao, S., Mohan, K., and Atreya, C. "Detection technologies for <i>Bacillus anthracis</i>" <i>Journal of Microbiological Methods</i> 2010. Volume 82. p. 747-748.] | ||
<sup> | <sup>14</sup>[http://www.ncbi.nlm.nih.gov/pubmed/6418708 Phillips, A.P., and Martin, K.L. "Comparison of direct and indirect immunoradiometric assays (IRMA) for <i>Bacillus anthracis</i> spores immobilised on multispot microscope slides". <i>Journal of Applied Bacteriology</i> 1983. Volume 41. p. 237-244.] | ||
<sup> | <sup>15</sup>[http://www.ncbi.nlm.nih.gov/pubmed/19793896 Kuehn A., Kovac, P., Saksena, R., Bannert, N., Klee, S.R., Ranisch, H., and Grunow, R. "Development of antibodies against anthrose tetrasaccharide for specific detection of <i>Bacillus anthracis</i> spores". <i>Clinical and Vaccine Immunology</i>. 2009. Volume 16. p. 1728-1737.] | ||
<sup> | <sup>16</sup>[http://www.ncbi.nlm.nih.gov/pubmed/11084608 Stopa, P.J. 2000. "The flow cytometry of <i>Bacillus anthracis</i> spores revisited". <i>Journal of Emergency Medicine</i>. 2000. Volume 41. p. 237-244.] | ||
<sup> | <sup>17</sup>[http://www.ncbi.nlm.nih.gov/pubmed/16283259 Edwards, K., Clancy, H., and Baeumner, A. "<i>Bacillus anthracis</i>:toxicology, epidemiology, and current rapid-detection methods." <i>Analytical and Bioanalytical Chemistry </i>. 2006. Volume 384. p. 73-74.] | ||
<sup> | <sup>18</sup>[http://aem.asm.org/content/67/8/3720 Qi, Y., Patra, G., Liang, X., Williams, L., Rose, S., Redkar, R., and DelVecchio, V. "Utilization of the <i>rpoB</i> gene as a specific chromosomal marker for real-time PCR detection of <i>Bacillus anthracis</i>". <i>Applied and Environmental Biology</i>. 2001. Volume 67. p. 3720-3727.] | ||
<sup> | <sup>19</sup>[http://wwwnc.cdc.gov/eid/article/8/10/02-0393_article.htm Hoffmaster, A.R., Meyer, R.F., Bowen, M.D., Marston, C.K., and Weyant, R.S., Thurman, K et al. "Evaluation and validation of a real-time polymerase chain reaction assay for rapid identification of <i>Bacillus anthracis</i>. "<i>Applied and Environmental Microbiology</i>". 2002. Volume 8. p. 1178-1182.] | ||
<sup> | <sup>20</sup>[http://www.ncbi.nlm.nih.gov/pubmed/12149348 Bell, C., Uhl, J., Hadfield, T., David, J., Meyer, R., Smith, T., and Cockerill, F. "Detection of <i>Bacillus anthracis</i> DNA by LightCycler PCR". <i>Journal of Clinical Microbiology</i>. 2002. Volume 40. p. 2897-2902.] | ||
<sup> | <sup>21</sup>[http://www.ncbi.nlm.nih.gov/pubmed/15522596 Wang, S.H., Wen, J.K., Zhou, Y.F., Zhang, Z.P., Yang, R.F., Zhang, J.B., Chen, J., and Zhang, X.E. "Identification and characterization of <i>Bacillus anthracis</i> by multiplex PCR on DNA chip". <i>Biosensors and Bioelectronics</i>. Volume 20. p. 807-813.] | ||
<sup> | <sup>22</sup>[http://www.ncbi.nlm.nih.gov/pubmed/12588551Ibkwe, A.M., Grieve, C.M. "Detection and quantification of <i>Escheria coli</i> O157: H7 in environmental samples by real-time PCR." <i>"Journal of Applied Microbiology</i>. 2003. Volume 94. p. 421-431.] | ||
Edited by Alison Lerner, a student of [http://www.jsd.claremont.edu/faculty/profile.asp?FacultyID=254/ Nora Sullivan] in BIOL187S (Microbial Life) in [http://www.jsd.claremont.edu/ The Keck Science Department of the Claremont Colleges] Spring 2013. | Edited by Alison Lerner, a student of [http://www.jsd.claremont.edu/faculty/profile.asp?FacultyID=254/ Nora Sullivan] in BIOL187S (Microbial Life) in [http://www.jsd.claremont.edu/ The Keck Science Department of the Claremont Colleges] Spring 2013. | ||
<!--Do not edit or remove this line-->[[Category:Pages edited by students of Nora Sullivan at the Claremont Colleges]] | <!--Do not edit or remove this line-->[[Category:Pages edited by students of Nora Sullivan at the Claremont Colleges]] | ||
Latest revision as of 04:42, 29 April 2013
Introduction
Bacillus anthracis is the causative agent of anthrax and has been adapted for use in bioterrorism. It is a large Gram-positive aerobic, rod shaped, bacillus bacterium (Figure 1). It ranges in size from 1-1.5 x 3-10 µm and is the only obligate pathogen within the genus bacillus.1 Anthrax is an acute disease, that appears suddenly and progresses rapidly, affecting humans and animals. Previously anthrax was perceived as a damaging disease affecting domesticated animals, the farming economy, and some industrial workers; however there has been a shift to now view the disease as an agent of bioterrorism. On October 5, 2001 the first anthrax related death in the United States occurred in over 25 years in which B. anthracis spores were mailed. 2 B. anthracis took the form of assymetric warfare where tactics, resources, and potential for destruction was unknown. According to the Center for Disease Control, approximately 1-2 cases of Anthrax occur each year; however in 2001, 22 cases of anthrax total occurred in the United States due to this act of bioterrorism (Figure 2).3
B. anthracis is successful as a bioterrorism agent due to its resistance to harsh environmental conditions through its ability to exist in a spore state. With the potential to have extreme and damaging effects on the population, the study of B. anthracis and the discovery of rapid and reliable technologies to identify the species is of utmost concern.
Biology and Genetics of B. anthracis
Virulence Plasmids
Two virulence plasmids, pXO1 and pXO2, encode the major virulence factors for B. anthracis.4 Plasmid pXO1 codes for three toxins which cause hemorrhage, edema, and necrosis and is 184.5 kilobase pairs (kbp) in size. The exotoxins it codes for are binary, which include the lethal factor (LF) and edema factor (EF), and the protein known as the protective antigen (PA). Therefore, the protective antigen it codes for acts as a binding domain on the host to permit entry of the toxin. pXO2 is a capsule bearing plasmid that is smaller than pXO1 having 95.3 kbp in size. It encodes three genes (cap A, cap B, and cap C) which are involved in the synthesis of the polyglutamyl capsule. The capsule prevents the bacteria from phagocytosis and is poorly recognized by the immune system of the host.5 Both pXO1 and pXO2 plasmids are necessary to provide the anthrax toxin and capsule respectively for B. anthracis to have full virulence. An attenuated strain results when either plasmid is lost.6
Three Proteins of Anthrax
The protective antigen (PA), lethal factor (LF), and edema factor (EF) are proteins that comprise the anthrax toxin. They are encoded by the genes pag, lef, and cya, respectively. The PA factor contains the cell-binding domain. LF or EF are responsible for cell toxicity by gaining access to the host cell's cytoplasms. This is able to occur when PA locates specific host cell surface receptors and cleavage by Furin, a cell surface protein, ensues. After cleavage, the remaining part of the PA auto-assembles in the cell membrane to form a heptameric (seven-membered) pore. The other protein components, EF or LF, then bind to the PA. PA enables the EF or LF toxins to then be released into the host cell's cytoplasm to cause infection (Figure 3). Once EF enters the cytoplasm, it raises cyclic adenosine monophosphate (cAMP) levels. This causes an inhibition of immune cell function, specifically resulting in edema, which is commonly referred to as swelling in which there is an accumulation of fluid trapped in a particular part of the body's tissues. If LF is released into the hosts' cells, it cleaves the hosts' protein kinases, which leads to altered signaling pathways and ultimately results in cell death. 7
Spore and Vegetative State
B. anthracis is a bacterium that can exist in both a spore and vegetative state, which make it a favorable agent for bioterrorism. A spore is a dormant and resilient structure that can exist for extended periods of time with a lack of nutrients. B. anthracis can form spores that are resilient to harsh environments, such as ultraviolet and ionizing radiation, heat, and various chemicals.8 A spore can germinate to exist in its vegetative state when the environmental conditions are favorable. To become a spore again, sporulation occurs, thereby ensuring the survival of B. anthracis during a destructive environmental condition.
Routes of Infection
Anthrax has three forms in which virulence occurs, which include cutaneous anthrax, gastrointestinal anthrax, and inhalational anthrax. When the host is exposed to the spores of B. anthracis via one of these three routes, infection occurs.
Cutaneous Anthrax
Cutaneous anthrax comprises over 90% of all human cases. It is obtained via a lesion on the skin in which infection occurs through an abrasion, cut, or insect bite.9 This type of anthrax manifests itself through a black eschar, which is dead tissue present on the surface of the skin as a scab, and is associated with edema. Some patients also experience symptoms of swollen lymph nodes, fever, and headache.
Gastrointestinal Anthrax
Gastrointestinal anthrax results from the ingestion of undercooked meat from animals who have B. anthracis in its vegetative form or dormant spore state. This form of the disease has a high mortality rate because it is difficult to make an early diagnosis due to its’ non specific presentation. Symptoms may include nausea, vomiting, appetite loss, fever, sore throat, and a swollen neck.
Inhalation Anthrax
Inhalation anthrax is another form in which anthrax can be contracted. This route of infection is particularly relevant for the contraction of anthrax when used as an agent of bioterrorism. Traditionally, inhalation anthrax was associated with industrial exposure as spores in the textile, meat packing, leather-tanning, bone meal processing, and hair/wool-sorting industries; however it became associated with bioterrorism in 2001 when anthrax was included in envelopes mailed within the United States. Beginning with “flu-like” symptoms of mild fever, fatigue, malaise (discomfort associated with illness or infection), myalgia (muscle pain), and non-productive cough from two to five days after initial exposure, the disease develops into an acute illness characterized by dyspnea (shortness of breath), stridor (wheezing caused by a narrowed or obstructed airway), fever, cyanosis (bluish discoloration of skin), and pleural effusion (excess fluid surrounding the lungs). With the initial display of "flu-like" symptoms when B. anthracis is inhaled, it difficult to diagnose the disease as anthrax. Anthrax manifests itself in the human body after inhalation of spores by first traveling to alveolar spaces within the lungs. Then the spores are transported through the lymphatic system to infect the entire body (Figure 4). Once the infection is established, it becomes refractory to treatment; therefore, the success of medical intervention is minimal. However, if the infection is diagnosed very early on, the disease may be successfully treated by multidrug antibiotic regimens and supported care; yet the prognosis of patients with inhalational anthrax is dismal as the mortality rate is high.10
Methods of Detection
A variety of methods exist for identifying B. anthracis. Phenotypic, antibody, and nucleic acid based methods are current techniques to identify the toxin in its various conditions including as a spore or in a vegetative state as well as using the microorganisms' DNA (Figure 5).
Conventional Phenotypic Methods of Identification
B. anthracis can be detected using conventional microbiological methods via phenotypic identification. The organism selectively grows on polymyxin-lysozyme EDTA-thallous acetate (PELT) agar.11 However, this method requires a 1-2 day incubation period and requires additional tests to confirm the presence of the disease. Additionally, B. anthracis can be visualized using McFadyean’s capsule-staining test which uses McFadyean’s polychrome methylene blue to stain the capsule and rods.12 Yet, these traditional methods for phenotypic identification of B. anthracis tend to be the most laborious and frequently require 1-2 days of testing. Laboratory personnel require additional training and certification to accurately identify this uncommon organism, which makes these methods impractical. Additionally, to confirm the presence of the pathogen, suspect isolates must frequently be further characterized through molecular techniques.13
Antibody Based Methods of Identification
Immunoradiometric assays, IRMA, utilizes an immunofluorescence system which acts to detect antigens displayed on the surface of a spore. This method can immobilize spores in order measure concentrations of antigens through the amount of antibody used. Typically, immunofluorescence assays provide a rapid detection of spores; however, specific serological detection is complex as a result of cross-reactivity between polyclonal and monoclonal antibodies and antigens as well as between B. anthracis and closely related species such as B. cereus. Despite these complications, the technique of immunoradiometric assays has been proven to successfully identify unique B. anthracis spore surface antigens. This was determined as a result of the lack of cross-reactivity with B. cereus, a closely related species.14 However, because the antibody must be radiolabeled, this method has been supplanted with the enzyme-linked immunosorbent assay.
The enzyme-linked immunosorbent assay, ELISA, detects immobilized spores of B. anthracis through identifying the spore surface antigens. Similar to an IRMA, an ELISA quantifies the amount of antibodies or proteins present in a sample; however unlike the IRMA, the ELISA method is a non-radioactive technique. The antigen must be immobilized when the ELISA is performed on a 96-well plate to permit high throughput results. For a successful ELISA, there must be a highly specific antibody-antigen interaction, which has been accomplished in the analysis of B. anthracis species. Antibodies displayed no cross-reactivity to other members of the B. cereus group, which is closely related to B. anthracis. Therefore, this technique is a reliable and effective technique for the identification of the B. anthracis species.15
Flow cytometry has also been used to detect B. anthracis spores through fluorescently labeled antibodies. A single wavelength of light is used to excite the fluorescent labels to disntiguish the entity of interest. The technique suspends cells in a stream of fluid and directs them through an electronic detection apparaturs. Flow cytometry is advantangeous because it allow the concurrent detection of multiple characteristics of particles. Quantum dots, which have narrower emission peaks than typical fluorophores used in flow cytometry, can be used in the detection B. anthracis. They result in an increase in accuracy of detecting cells in real-time and over extended periods of time. The quantum dots cause an increase in brightness and report more consistent results than traditional fluorophores. Although flow cytometry is a rapid method, it has a low limit of detection of 103 spores, therefore samples must be in abundance, which is not ideal.16
Nucleic Acid Based Methods of Identification
Nucleic acid based methods, including PCR, Real-Time PCR, and multiplex PCR, for the detection of B. anthracis rely on the selection of nucleic acid probes that are specific to the species and must have no cross-reactivity with other species. Primers frequently target amplification of the genes, lef, pag, and cap located on either or both of the virulence plasmids pXO1 and pXO2. 17
Using PCR, strains containing the plasmids pXO1 and pXO2 are amplified. Analysis by gel electrophoresis follows as well as DNA sequencing. These methods can provide a measure of the genetic variability between isolates. The sequence rpoB is commonly used in PCR amplification of B. anthracis. The 175 base-pair sequence is highly selective for B. anthracis because it has the ability to discriminate B. anthracis from other Bacillus strains. Only one strain out of 175 tested bacilli strains has been found to be cross-reactive.18 Ultimately, PCR is able to detect the presence of both virulence plasmids, can characterize B. anthracis vaccine strains, and detect spores from soil samples.
Real-Time PCR, also known as quantitative PCR (qPCR), utilizes fluorescence to detect the DNA it amplifies. Fluorescence increases in a real-time manner as additional copies of DNA are produced. This type of assay avoids the need for further processing of the DNA to detect and quantify amplified sequences separately as necessary with PCR. Additionally, amplification is observed in real-time rather than after the fact. Since amplification and detection occur in the same chamber, there is a reduced likelihood for cross-contamination of products. In the 2001 anthrax outbreak in the United States in which B. anthracis was used as an agent of bioterrorism, real-time PCR was used to test and compare 542 isolates from clinical, environmental, and suspect powder samples from both this outbreak and prior outbreaks.19 Every hour, five copies per real-time PCR reaction of B. anthracis was detected and amplified. Before amplification occurred, the method required a growth period for 18-24 hours and an extraction time of 0.5-1 hour.20 100% specificity was reported using this type of assay even when 56 non B. anthracis species were tested.
Multiplex PCR amplifies specific regions of DNA using numerous sets of primers in a single reaction tube. By amplifying and detecting multiple target sequences on DNA, multiplex PCR is beneficial in eliminating false-negative and false-positive results.21
These amplification methods are often combined with high-throughput techniques for sample preparation and detection. Nucleic acid based systems are more sensitive than antigen and antibody-based detection systems in that they have the ability to detect fewer microorganisms in shorter amounts of time.22 Each method of PCR amplification has their own unique benefits for amplifying DNA; however, not all techniques are ideal to detect low levels of bacteria in environmental samples. For example, the downfall of real-time PCR is that this technique requires a DNAse step to remove DNA to prevent amplification of both DNA and RNA. The DNAse step to remove DNA has been proven to not be completely effective; therefore additional complex and time-consuming steps are necessary to ensure accurate results.
Conclusion
B. anthracis has many genetic similarities among various Bacillus species and can exist in the spore and vegetative state making it an ideal agent for bioterrorism. These features make the detection and specific identification of B. anthracis difficult and have proven to require complex techniques and laborious methods. However, targeting the virulence plasmids pXO1 and pXO2 of B. anthracis have successfully been used in detecting the species. As an agent for a potential biological threat, B. anthracis is more frequently being studied. Due to the heightened interest in the organism, there have been a large number of detection technologies that have recently been developed and are underdevelopment to reliably and quickly characterize and identify the species with the hopes of controlling anthrax outbreaks and treating the outbreaks rapidly.
References
2Spencer, R.C. "Bacillus anthracis". Journal of Clinical Pathology. 2003. Volume 56. p. 182-187.
3"Questions and answers about Anthrax". Center for Disease Control and Prevention.. 2012.
6Spencer, R.C. "Bacillus anthracis". Journal of Clinical Pathology. 2003. Volume 56. 182-187.
Edited by Alison Lerner, a student of Nora Sullivan in BIOL187S (Microbial Life) in The Keck Science Department of the Claremont Colleges Spring 2013.
