Virulence of Pseudomonas savastanoi: Difference between revisions
No edit summary |
|||
| (62 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
<br><b>Hildy Joseph, Kenyon 2013</b> | <br><b>Hildy Joseph, Kenyon 2013</b> | ||
==<b><font size="5">Introduction</font></b>== | ==<b><font size="5">Introduction</font></b>== | ||
[[Image:Olive.jpg|thumb| | [[Image:Olive.jpg|thumb|150px|left|Fig 1. Olive knot disease infecting an olive plant. Source: http://www.cbgp.upm.es/en/phytopathogenic_bacteria.php]] | ||
<br>Olive knot disease, which is characterized by knots and galls on emerging stems, branches, and occasionally leaves, was first described in the fourth century B.C. by the Greek philosopher Teophrastus. [[Image:Teophrastus.jpg|thumb| | <br>Olive knot disease, which is characterized by knots and galls on emerging stems, branches, and occasionally leaves [Fig 1.], was first described in the fourth century B.C. by the Greek philosopher Teophrastus [Fig. 2]. [[Image:Teophrastus.jpg|thumb|150px|right|Fig 2., the Greek philosopher to first document olive knot disease in the fourth century B.C. Source: http://www.biografiasyvidas.com/biografia/t/teofrasto.htm]] <i>Pseudomonas savastanoi</i> is the bacteria responsible for the condition [1]. Infection occurs through wounds incurred by pruning or from severe weather events like freezing and hail [2]. A wide range of symptoms can result from infection, including death and decay. Other consequences of the pathogen include defoliation, reduced limb and overall plant size. Infections can be asymptomatic for up to eight months from exposure [1]. Knots usually form in the spring during the most active growth period [2]. Plant vulnerability is also increased in the autumn, in part because of other pathogens like the fungus <i>Spilocaea oleaginea</i> target olive trees concurrently. Thus, the chance of co-infection by <i>P. savastanoi</i> is heightened [4]. The bacterium spreads easily within an orchard via wind and rain as well as pruning tools [2]. Moreover, once infected, <i>P. savastanoi</i> can spread to new wound sites within the same plant [5]. Two cultivars were both inoculated with <i>P. savastanoi</i> to determine its spread through orchards. All trees, regardless of inoculation, were prone to infection. However, inoculated trees were significantly less vigorous over the four-year study [Fig. 3] [3]. [[Image:Wiki fig.jpg|thumb|300px|right|Fig. 3Two cultivars of olive trees were inoculated with <i>P. savastanoi</i> and then compared with non-inoculated trees over four years [http://onlinelibrary.wiley.com/store/10.1111/j.1365-3059.2009.02200.x/asset/j.1365-3059.2009.02200.x.pdf?v=1&t=hghzk7oo&s=3ac81d8a2788062380e5226dec3f87bc3fd0db9d "[3]"]]] Trees that were not inoculated tended to have better health, in general [3]. Olive quality from infected plants is greatly reduced. Olives from infected plants are bitter, salty, sour, or rancid [1]. Yield is also negatively impacted by the disease. Olive knot disease is a global issue and has been found in parts of Europe, Asia, Africa, North America, South America, and Australia. The incidence and degree of virulence varies by location [2]. | ||
<br>< | <br>Understanding the virulence of <i>P. savastanoi</i> is important because of the emerging economic significance of olives and olive oil [Fig. 4]. [[Image:Olive-oil-and-mature-olives.-.jpg|thumb|150px|right|Fig. 4 Promoting olive crops has been of interest due to the proven health benefits of the Mediterranean diet, which includes olives and olive oil. Source: http://www.walksofitaly.com]] Olive oil comprises a major part of the Mediterranean diet, which is becoming increasingly popular because of the associated health benefits. The Mediterranean diet may contribute to reducing LDL-cholesterol and improving HDL-cholesterol, which lessens the risk for heart disease. Digestion, autoimmune, and inflammatory diseases can all also be improved with the Mediterranean diet [6] [7] . <i>P. savastanoi</i> infection is part of a greater environmental concern about the high incidence of monoculture in crop orchards. Reducing genetic diversity can increase susceptibility to disease. Thus, understanding plant pathogens is essential for preserving the world's food supplies [8]. | ||
<br> | ===<br><b><font size="4">Classification of P. savastanoi</font></b>=== | ||
< | |||
<br>< | <br>The genus <i>Psuedomonas</i> consists of Gram-negative bacteria that are highly prevalent in nature. Both phytopathogenic as well as nonvirulent species are part of the group, in addition to the single human pathogen, <i>Pseudomonas aeruginosa</i> [9] [10] [11]. <i>P. savastonoi</i> is a nonsporeforming bacteria that contains several flagella for motility [9]. The occurrence of phytopathogeny is polphyletic among species of Pseudomonas, which provides evidence for horizontal gene transfer of virulence genes. The virulence genes are termed the '<i>P. syringae</i> complex' and may have first arisen in the common ancestor for the clade of plant pathogens within <i>Pseudomonas</i>, which includes <i>P. savastanoi</i>. More recently evolutionarily, the virulence genes may have been transferred horizontally to other species in the <i>Pseudomonas</i> genus. The complex comprises a pathogenicity island. [10]. Generally, pathogens in the <i>P. syringae</i> complex cause leaf death and cankers, and thus the olive knots caused by <i>P. savastanoi</i> pv. savastanoi are unique [12]. | ||
<br> | <br>P. savastonoi was originally thought to be one of forty-five pathovars of <i>Pseudomonas syringae</i> due to similarities in biochemistry and physiology. DNA-DNA hybridization data revealed that <i>Pseudomonas savastanoi</i> actually has sufficient genetic distinction from <i>P. syringae</i> to merit its own species classification. Several pathovars were subsequently attributed to P. savastanoi, including <i>P. savastanoi</i> pv. savastanoi, <i>P. savastanoi</i> pv. glycinea, and <i>P. savastanoi</i> pv. phaseolicola [13]. Pathovars that have subsequently been recognized include <i>P. savastanoi</i> pv. fanxini and <i>P. savastanoi</i> pv. nerii. | ||
== | ===<br><b><font size="4">Host range and susceptibility</font></b>=== | ||
<br><b><font size="4"> | |||
<br> | <br>Both cultivated and wild olives (<i>Olea europaea</i>) as well as ash (<i>Fraxinus excelsior</i>) are susceptible to tumorigenesis as a result of <i>P. savastanoi</i> pv. savastanoi. There is some overlap in hosts with other closely related pathovars. <i>P. savastanoi</i> pv. nerii infects oleander (<i>Nerium oleander</i>) but can also infect olives. <i>P. savastanoi</i> pv. savastanoi has been shown to infect oleander, but the occurrence is unusual [12]. Though <i>P. savastanoi</i> pv. savastanoi is generally limited to olive and oleander hosts, there is a recent report of a <i>P. savastanoi</i> infection in Mandevilla sanderi, commonly known as Brazilian jasmine [14]. There are several other pathogenic pathovars in the <i>P. savastanoi</i> species. <i>P. savastanoi</i> pv. glycinea infects soybeans and <i>P. savastanoi</i> pv. phaseolicola is responsible for halo blight in beans [13]. | ||
[ | |||
<br> | <br>Resistant olive cultivars are rare, though the amount susceptibility to <i>P. savastanoi</i> does vary. Young olive plants are particularly non-resistant to infection [5]. Interestingly, <i>P. savastanoi</i> can be epiphytic upon infection [15]. | ||
==<font size="5"><b>Mechanisms of Virulence</b></font>== | |||
===<br><b><font size="4">Plant hormones: cytokinin and IAA</font></b>=== | |||
<br> | |||
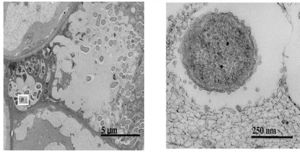
<br>[[Image:9688365.jpg|thumb|300px|right|Fig. 5 Scanning electron micrograph of olive knot. Shows <i>P. savastanoi</i> bacteria infecting host cell. [http://www.ncbi.nlm.nih.gov/pubmed/21255279 "[15]"]]] Infection by <i>Pseudomonas savastanoi</i> causes cavities in host cells in which bacteria can aggregate [Fig. 5] [16]. <i>P. savastanoi</i> also accumulate in the xylem, which suggests a mechanism, heretofore unverified, of bacterial transport throughout the plant [15]. | |||
<br>Plant hormones are one aspect of virulence that activates tumorigenesis. Cytokinin and IAA production both contribute to knot formation by interfering with endogenous signals to induce rapid proliferation of host cells at the site of infection [16] [17] [18]. <i>Pseudomonas savastanoi</i> carry plasmids that encode for indole-3-acetic acid (IAA), a protein involved in auxin signaling [17] [19]. Various genetic studies confirm the importance of IAA and cytokinins for virulence. Mutants lacking either IAA or cytokinins were less symptomatic upon infection of a host olive plant. Without IAA or cytokinins, infections were limited to displaying only some of the normal symptoms, such as leaf necrosis and stem swelling. IAA is sufficient for tumor formation, though cytokinins also play an important role [20]. Strains lacking the pIAA1 plasmid, which encodes for the two genes responsible for IAA production, iaaM and iaaH, fail to induce olive knot growth in hosts plants [21]. Auxin may be important for virulence by downregulating factors that contribute to the host defense. For example, auxin can protect against the hypersensitive response, a defense mechanism in plants injected with <i>P. syringae</i> [22]. HR (hypersensitive response) is a form of programmed cell death in host cells and is widely conserved among plants and animals [23]. Auxin has many additional roles. It may regulate gene expression or affect the physiology of host cells, for example [21]. | |||
<br><i>P. savastanoi</i> occur in conjunction with other bacterial species within olive knots. It is hypothesized that phytohormones produced by the nonpathogenic bacteria coexisting in olive knots may influence <i>P. savastanoi</i> [12]. Auxin is produced by a variety of other genera and species that are common to olive knots, including <i>Stenotrophomonas maltophila</i>, <i>Pantoea agglomerans</i>, <i>Pantoea oleae</i>, <i>Burkholderia cepacia</i>, and <i>Hafrnia alvei</i>, all of which are other Gram-negative rod forming bacteria. Furthermore, IAA specifically is produced by other </i>Pseudomonas</i> species [24]. | |||
===<br><b><font size="4">Suppressing host immune response and defense mechanisms</font></b>=== | |||
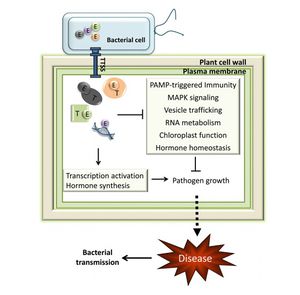
<br>The <i>P. Syringae</i> complex, which is made up of the <i>hrp</i>/<i>hrc</i> gene cluster, acts on the host immune system [18] [23]. The <i>hrp</i> genes encode for a type III protein secretion system (T3SS) [Fig. 6]. The T3SS is comprised of a pilus that connects the bacterium with the host membrane and the pore that forms in the host cell to allow entry of type III effector proteins. All of the proteins associated with the T3SS are encoded for in the <i>P. Syringae</i> pathogenicity island [23]. T3SS are found in Gram-negative bacteria that interact eukaryotes, both as pathogens or symbionts, such as nitrogen-fixing <i>Rhizobium</i> [25]. General defense mechanisms in plants involve mechanical resistance to pathogens through the cell wall. Additionally, plants contain a set of receptors that respond to microbe-associated molecular patterns to induce production of antimicrobial agents [23]. The T3SS complex can overcome plant cell walls with the aid of helper proteins that are secreted into the extracellular space [23]. Host cell wall degradation has been observed in in olive knots [15]. | |||
<br>The T3SS proteins evolved from a common ancestor and the genomic island encoding all of the associated proteins was transferred horizontally among seven families. <i>P. savastanoi</i> and other Pseudomonads exhibit the <i>Hrc-Hrp2</i>-T3SS family. The other genera that share T3SS-2 are <i>Erwinia</i>, <i>Ralstonia</i>, and <i>Xanthomonas</i> [25]. | |||
[[Image:TTSS.jpg|thumb|300px|left|Fig. 6 General mechanism of Type III Secretion System (T3SS) in plant pathogens. [21]]] | |||
One emerging area of research is understanding the specific effectors and other elements of the T3SS [26]. The gene clusters are essential to <i>P. savastanoi</i> virulence and may be more important pathologically than auxin and cytokinin action. The <i>hrp</i> gene cluster reduces plant vitality by inducing leakage of essential nutrients into the extracellular milieu, in addition to its roles inhibiting the plant immune system. The closely related <i>hrc</i> genes are responsible for the plant HR [18]. | |||
<br>In addition to the T3SS, other interactions with host defense mechanisms have been postulated. Plants produce a range of secondary metabolites that aid in defense. Flavonoids are one class of phenolic compounds that are protective against parasites and pathogens [27]. Flavonoids, along with other phenolic compounds like secoiridoids and lignans are common to olive oil and confer some of the major health benefits associated with the Mediterranean diet [7] [28]. Sequence data and empirical evidence in <i>P. savastanoi</i> reveals the potential for phenol catabolism. Other pathogens of woody plants, in which phenolic defense mechanisms are common, show similar catabolic abilities [26]. Furthermore, olive knots induced by <i>P. savastanoi</i> pv. savastanoi show a higher presence of phenolic compounds than in other leafs and shoots [29]. Thus, <i>P. savastanoi</i> may circumvent the the secondary metabolites produced by plants to preempt and actively combat bacterial infections by catabolizing the compounds [26]. | |||
===<br><b><font size="4">Emerging views of virulence: signaling mechanisms and outer membrane vesicles</font></b>=== | |||
[[Image:Hildy pic 1.jpg|thumb|300px|right|Fig. 7 Visualization of outer membrane vesicles, which help facilitate P. savastanoi pathogenicity. (left)Knot tissue invaded by P. savastanoi microbes. (right) Release of outer membrane vesicles forming from host cell. Modified from [30]]] | |||
<br>The <i>P. savastanoi</i> signaling mechanism is another quality that has been implicated in its virulence. Gram-negative bacteria commonly use the N-acyl homoserine lactone (AHL) signaling molecules. <i>P. savastanoi</i> often occurs in association with <i>P. agglomerans</i> and <i>E. toletana</i>, which are both nonpathogenic Enterobacteriaceae that frequently grow at olive knot infection sites [31]. In general, plants that have been inoculated with <i>P. savastanoi</i> have a higher overall bacteria population than plants that do not bear an infection by <i>P. savastanoi</i> [3]. AHL signals can be shared across <i>P. agglomerans</i>, <i>E. toletana</i>, and <i>P. savastanoi</i>. Specifically, the association of <i>E. toletana</i> and <i>P. savastanoi</i> confers synergistic pathogenic effects. The interactions between <i>E. toletana</i> and <i>P. savastanoi</i> are stable and lead to a higher rate of olive knot formation [31]. Alternatively, interactions between <i>P. savastanoi</i> and resident microbial communities on host plants may negatively impact bacterial virulence [31] [32]. The pathogenicity of <i>P. Syringae</i> pv. syringae, which is closely related species to <i>P. savastanoi</i> pathovars, was reduced by crosstalk with signaling molecules from the resident bacterial populations. Instead of synergistic effects, the AHLs from other species interfered with AHL signaling in <i>P. Syringae</i> [32]. Researchers are working on exploiting the interactions between resident populations and <i>P. savastanoi</i> to reduce its incidence and virulence [33]. | |||
<br>Additionally, <i>P. savastanoi</i> employ a unique mechanism among plant pathogens by inducing the release of outer membrane vesicles from the host cell [Fig. 7]. Animal pathogens commonly release outer membrane vesicles for transporting virulence factors into host cells. The role of the membrane vesicles in a <i>P. savastanoi</i> infection may be a relating to correcting for osmotic pressure [30]. | |||
===<br><b><font size="4">Development of Genetic Tools for Understanding Virulence</font></b>=== | |||
<br>Major advances in the last several years have rapidly progressed the understanding of <i>P. savastanoi</i> after decades of stalled research. Before 2007, the significant impediment to studying <i>P. savastanoi</i> was the lack of any strains that were amenable to genetic manipulation, specifically conjugation. The problem is consistent across other Pseudomonads that infect woody plants, including <i>P. savastanoi</i> pvs. Savastanoi, franxini, and nerii. Several strains were isolated that are have more efficient transformations [34]. One of the strains with the highest transformation efficiency, <i>P. savastanoi</i> pv. savastanoi NCPPB 3335, was recently sequenced [26] [34]. A draft genome was obtained with pyrosequencing and compared to other published <i>P. Syringae</i> genomes. The primary goal of the genetic manipulation is to augment the current poor understanding of <i>P. savastanoi</i> virulence [26]. | |||
<br>The draft genome of <i>P. savastanoi</i> pv. savastanoi NCPPB 3335 revealed 73 genes that are not found in other species as well as eight gene inversions in comparison with closely related species. Researchers suggest that the basis for host specificity is encoded for in the 73 genes unique to <i>P. savastanoi</i> [26] [35]. | |||
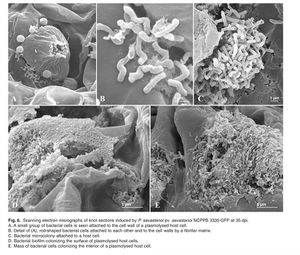
<br>Other emerging techniques have recently allowed for easier plant manipulation and greater resolution. For example, cultivation by micropropagation circumvents the arduous growing requirements for olive trees, including long growing seasons and large plant size [Fig. 8]. Additionally, the draft <i>P. savastanoi</i> pv. savastanoi NCPPB 3335 genome has been employed to develop a green fluorescent protein tagging system. Scanning and transmission electron microscopy can be used to visualize the bacteria in planta and further elucidate virulence mechanisms [12]. [[Image:microprop.jpg|thumb|150px|left|Fig 8. An example of micropropagation with strawberry plants. The method is efficient with both space and time. Source: http://crazygardeners1.wordpress.com/micropropagation/]] | |||
==<b><font size="5">Responding to Olive Knot Disease in Crop Management</font></b>== | |||
===<br><b><font size="4">Detection of P. Savastanoi</font></b>=== | |||
<br>An infection can easily be recognized on crop plants in its late stages. However, insect wounds are easily confused with olive knots in the preliminary onset. Bacteria can be isolated from knots and cultured in the laboratory for identification and confirmation, though older knots generally contain a variety of secondary invaders [2]. Moreover, plants can be infected without having visible symptoms [36]. Furthermore, <i>P. savastanoi</i> grows relatively slowly in the laboratory, with a required incubation between two to three days at 27°C [2] [9]. Other techniques have been developed to rapidly detect <i>P. savastanoi</i> without culturing or the necessity of serological methods. For example, there is a highly sensitive PCR technique based on amplification of the iaaL gene, which encodes for a product that catalyzes the addition of lysine to IAA [36]. Isolates can be distinguished by sequence differences, specifically differing numbers of nucleotide motif tandem repeats, in both iaaL paralogs found in different <i>P. savastanoi</i> strains [37]. Another technique using rt-PCR can simultaneously detect <i>P. savastanoi</i> along with four RNA viruses from a single host sample. Detecting multiple plant pathogens in a single reaction conserves time and resources [6]. | |||
===<br><b><font size="4">Control of the pathogen</font></b>=== | |||
<br>Farmers have a variety of options to alleviate the problem of olive knot disease, though noncommercial farmers have fewer available products. Landscapers and homeowners are advised to practice several strategic planting practices to minimize the chances for disease dissemination. For example, timing of olive tree purchase is critical, as <i>P. savastanoi</i> is often epiphytic during the winter months and infections are most visible in late spring. Furthermore, visible galls should be removed with sterile pruning tools to minimize spread of the bacteria [38]. The levels of epiphytic bacteria may be positively correlated with the occurrence of olive knots, and thus minimizing the presence of olive knots reduces the overall spread of the infection [39]. One measure to preemptively combat disease is to remove oleander plants that are growing in close proximity to growing area because of the potential overlap in host specificity for <i>P. savastanoi</i> [38]. However, <i>P. savastanoi</i> is generally an obligate symbiont and does not proliferate on soil [12]. | |||
<br>Resistant olive cultivars would be the best method for controlling P. savastanoi. However, naturally resistant cultivars have not yet been detected. Recently, susceptibility measures to <i>P. savastanoi</i> have been standardized, which will aid in developing cultivars that will not become infected [12]. | |||
====<br><b><i>Chemical</i></b>==== | |||
<br>Copper compounds are commonly used to reduce <i>P. savastanoi</i> proliferation [33]. The Cu<sup>2+</sup> ions can be toxic to P. savastanoi. Copper can also induce a viable but nonculturable state, which interferes with serological detection of <i>P. savastanoi</i> on host plants [39]. The method poses threats to the environment and public health. Furthermore, control of <i>P. savastanoi</i> with copper shows reduced efficacy with continued use [33]. A couple copper compounds are available, including copper oxychloride and cuprocalcic suflate plus mancozeb, which have been shown to be equally effective in reducing the amount of <i>P. savastanoi</i> growing on the leafs and stems [39]. Maximal efficacy of treatment requires a minimum of biannual application in the spring and fall, when olive trees are at the highest risk for infection. Olive trees are predisposed for infection when leafs fall in the spring due to residual leaf scarring as well as during frosts in the late autumn [4]. Bacteria that resist the copper treatment and exist in epiphytic and endophytic states can proliferate with failure to adhere to a regular copper treatment schedule [39]. However, farmers are recalcitrant toward biannual copper application because of its effect on the olive product [4]. | |||
<br>Polyphenols have also demonstrated efficacy in controlling olive knot. The compounds were obtained in extracts of olive mill waste water. Even at the lowest concentration tested, 100 mg/L, all olive knot formation was blocked, whereas a concurrent copper treatment was less effective [33]. | |||
====<br><b><i>Biological</i></b>==== | |||
<br>Other bacterial species can be used to control the incidence of <i>P. savastanoi</i>. Antimicrobial proteins produced by other microbes include colicin-like bacteriocins. Bacteriocins are common bacterial protein products that are detrimental to closely related species. Bacteriocins from <i>P. Syringae</i> pv. ciccaronei can inhibit both olive knot growth as well as epiphytic P. savastoni infection [8]. Another promising development in the biological control of olive blot is the application of Bacillus subtilis. B. subtilis produces antibiotics like zwittermicin-A and kanosamine as well as other antimicrobial lipoproteins. A protein produced from B. subtilis has been shown to reduce the size of olive knots caused by P. savastanoi [40]. | |||
==<b><font size="5">Conclusion</font></b>== | |||
Olive knot disease is of great economic importance due to the increasing interest in the benefits of polyunsaturated fatty acids in the diet. Farmers maximize profit and minimize the necessity of environmentally harmful treatments if pests can be controlled. <i>P. savastanoi</i> is the major threat to olive trees. Exciting breakthroughs have occurred in recent years relating to the genetic manipulation of <i>P. savastanoi</i>, which will allow for a greater understanding of the plant pathogen's virulence. | |||
==<font size="5"><b>References</b></font>== | |||
[1] Iacobellis NS. 2001. Olive knot. In: Maloy OC, Murray TD, eds. Encyclopedia of plant pathology. New York, NY, USA: John Wiley & Sons, 7143–715. | |||
<br>[2] [http://link.springer.com/article/10.1071/AP03074 "Young, J.M. 2004. Olive knot and its pathogens. Australian Plant Pathology 33:33-39."] | |||
<br>[3] [http://onlinelibrary.wiley.com/store/10.1111/j.1365-3059.2009.02200.x/asset/j.1365-3059.2009.02200.x.pdf?v=1&t=hghzk7oo&s=3ac81d8a2788062380e5226dec3f87bc3fd0db9d "Quesada, J.M., R. Penyalver, J. Perez-Panades, C.I. Salcedo, E.A. Carbonell, M.M. Lopez. 2009. Dissemination in Pseudomonas savastanoi pv. savastanoi populations and subsequent appearance of olive knot disease. Plant Pathology 59(2):262-269."] | |||
<br>[4] [http://apsjournals.apsnet.org/doi/pdfplus/10.1094/PDIS.2004.88.2.131 "Teviotdale, B.L., W.H. Krueger. 2004. Effects of copper sprays, defoliation, rainfall, and inoculum concentration on incidence of olive knot disease. Plant Disease 88:131-135."] | |||
<br>[5] [http://apsjournals.apsnet.org/doi/pdf/10.1094/PHYTO-96-0313 "Penyalver, R., A. Garcia, A. Ferrer, E. Bertolini, J.M. Quesada, C.I. Salcedo, J. Piquer, J. Perez-Panades, E.A. Carbonell, C. del Rio, J.M. Caballero, M.M. Lopez. 2006. Factors affecting Pseudomonas savastanoi pv. savastanoi plant inoculaitons and their use for evaluation of olive cultivar susceptibility. Phytopathology 96(3):313-319."] | |||
<br> [6] [http://apsjournals.apsnet.org/doi/pdfplus/10.1094/PHYTO.2003.93.3.286 "Bertolini, E., A. Olmos, M.M. Lopez, M. Cambra. 2003. Multiplex nested reverse transcription-polymerase chain reaction in a single tube for sensitive and simultaneous detection of four RNA viruses and Psuedomonas savastanoi pv. savastanoi in olive trees. Phytopathology 93(3):286-292."] | |||
<br> [7] [http://nutricore.com.br/app/webroot/img/bibliotecas/Dieta%20Mediterranea%20e%20Saude%20-%20impacto%20do%20azeite.pdf "Alarcon de la Lastra, C., M.D. Barranco, V. Motilva, J.M. Herrerias. 2001. Mediterranean diet and health: biological importance of olive oil. Current Pharmaceutical Design 7(10):933-950."] | |||
<br> [8] [http://www.ncbi.nlm.nih.gov/pubmed/23176505 "Grinter, R., J. Milner, D. Walker. 2012. Bacteriocins active against plant pathogenic bacteria. Biochemical Society Transactions 40(6):1498-1502."] | |||
<br> [9] [http://ijs.sgmjournals.org/content/32/2/166.full.pdf+html "Janse, J.D. 1982. Pseudomonas syringae subsp. Savastanoi (ex Smith) subsp. Nov., nom. Rev., the bacterium casuing excrescences on Oleaceae and Merium oleander L. International Journal of Systemic and Evolutionary Microbiology 32(2):166-169."] | |||
<br> [10] [http://mic.sgmjournals.org/content/146/10/2385.full.pdf+html "Yamamoto, S., H. Kasai, D.L. Arnold, R.W. Jackson, A. Vivian, S. Harayama. 2000. Phylogeny of the genus Psuedomonas: intrageneric strucure reconstructed from the nucleotide sequences of gryB and rpoD genes."] | |||
<br> [11] [http://cid.oxfordjournals.org/content/27/Supplement_1/S93.long "Hancock, R.E.W. 1998. Resistance mechansims in Psuedomonas aeruginosa and other nonfermentative gram-negative bacteria. Clinical Infectious Diseases 27(S1):S93-S99."] | |||
<br> [12] [http://onlinelibrary.wiley.com/doi/10.1111/j.1364-3703.2012.00816.x/full "Ramos, C., I.M Matas, L. Bardaji, I.M. Argon, J. Murillo. 2012 Pseudomonas savastanoi pv. savastanoi: some like it knot. Molecular Plant Pathology 13(9):998-1009."] | |||
<br> [13] [http://ijs.sgmjournals.org/content/42/4/606.full.pdf "Gardan, L., C. Bollet, M. Abu Ghorrah, F. Grimont, P.A.D. Grimont. 1992. DNA relatedness among the pathovar strains of Pseudomonas syringae subsp. Savastanoi Janse (1982) and proposal of Psuedomonas savastanoi sp. nov. International Journal of Systemic and Evolutionary Microbiology 42(4):606-612."] | |||
<br> [14] [http://www.ncbi.nlm.nih.gov/pubmed/23023746 “Eltlbany, N., Z.Z. Prokscha, M.P. Castaneda-Ojeda, E. Krogerrecklenfort, H. Heuer, W. Wohanka, C. Ramos, K. Smalla. 2012. A new bacterial disease on Mandevilla sanderi, caused by Pseudmonas savastanoi: lessons learned for bacterial diversity studies. Applied Environmental Microbiology 78(23):8492-8297.”] | |||
<br> [15] [http://www.ncbi.nlm.nih.gov/pubmed/21255279 “Rodriguez-Moreno, L., A.J. Jimenez, C. Ramos. 2009. Endopathogenic lifestyle of Pseudomonas savastanoi pv. Savastanoi in olive knots. Microbial Biotechnology 2(4):476-488.”] | |||
<br>[16] [http://onlinelibrary.wiley.com/doi/10.1111/j.1365-3059.2008.01935.x/pdf “Marchi, G., B. Mori, P. Pollacci, M. Mencuccini, G. Surico. 2009. Systemic spread of Pseudomonas savastanoi pv. Savastanoi in olive explants. Plant Pathology 58:152-158.”] | |||
<br>[17] [http://www.sciencedirect.com/science/article/pii/0048405985900062 “Surico, G., N.S. Iacobellis, A. Sisto. 1985. Studies on the role of indole-3-acetic acid and cytokinins in the formation of knots on olive and oleander plants by Pseudomonas syringae pv. Savastanoi. Physiological Plant Pathology 26:309-320.”] | |||
<br>[18] [http://apsjournals.apsnet.org/doi/pdfplus/10.1094/PHYTO.2004.94.5.484 “Sisto, A., M.G. Cipriani, M. Morea. 2004. Knot formation caused by Pseudomonas syringae subsp. savastanoi on olives is hrp-dependent. Phytopathology 94(5):484-489.”] | |||
<br>[19] [http://www.landesbioscience.com/journals/psb/MutoPSB2-5.pdf “Muto, H., M.K. Watahiki, K.T. Yamamoto. 2007. What makes each Aux/IAA gene unique in its gene family, expression pattern or properties of the gene product? Plant Signaling and Behavior. 2(5):390-392.”] | |||
<br>[20] [http://onlinelibrary.wiley.com/doi/10.1111/j.1439-0434.1994.tb04813.x/abstract “Iacobellis, N.S., A. Sisto, G. Sucrico, A. Evidente, E. DiMaio. 1994. Pathogenicity of Pseudomonas syringae subsp. savastanoi mutants defective in phytohormone production. Journal of Phytopathology 140(3):238-248.”] | |||
<br>[21] [http://cshperspectives.cshlp.org/content/3/4/a001438.full.pdf+html “Spaepen, S., J. Vanderleyden. 2010. Auxin and plant-microbe interactions. Cold Spring Harbor Perspectives in Biology 3(4):a001438.”] | |||
<br>[22] [http://link.springer.com/content/pdf/10.1007%2F978-3-642-30141-4_62.pdf “Melotto, M., B.N. Kunkel. 2013. Virulence strategies of plant pathogenic bacteria. In: The Prokaryotes – Prokaryotic Physiology and Biochemistry, eds. E. Rosenberg et al. Springer-Verlag, Berlin, pp. 61-82.”] | |||
<br>[23] [http://www.annualreviews.org/doi/pdf/10.1146/annurev.micro.60.080805.142251 “Grant, S.R., E.J. Fisher, J.H. Chang, B.M. Mole, J.L. Dangl. 2006. Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annual Reviews of Microbiology 60:425-449.”] | |||
<br>[24] [http://onlinelibrary.wiley.com/doi/10.1002/jobm.200800036/pdf “Ouzari, H., A. Khsairi, N. Raddadi, L. Jaoua, A. Hassen, M. Zarrouck, D. Daffonchio, A. Boudabous. 2008. Diversity of auxin-producing bacteria associated to Pseudomonas savastanoi-induced olive knots. Journal of Basic Microbiology 48:370-377.”] | |||
<br>[25] [http://www.biomedcentral.com/1471-2180/12/188 “Gazi, A.D., P.F. Sarris, V.E. Fadouloglou, S.N. Charova, N. Mathioudakis, N.J. Panopoulos, M. Kokkinidis. 2012. Phylogenetic analysis of a gene cluster encoding an additional, rhizobial-like type III secretion system that is narrowly distributed among Pseudomonas syringae strains. BMC Microbiology 12:188.”] | |||
<br>[26] [http://onlinelibrary.wiley.com/doi/10.1111/j.1462-2920.2010.02207.x/full “Rodriguez-Palenzuela, P., I.M. Matas, J. Murillo, E. Lopez-Solanilla, L. Bardaji, I. Perez-Martinez, M.E. Rodriguez-Moskera, R. Penyalver, M.M. Lopez, J.M. 2010. Annotation and overview of the Pseudomonas savastanoi pv. savastanoi NCPPB 3335 draft genome reveals the virulence gene complement of a tumour-inducing pathogen of woody hosts. Environmental Microbiology 12(6):1604-1620.”] | |||
<br>[27] [http://onlinelibrary.wiley.com/doi/10.1055/s-2005-873009/pdf “Treutter, D. 2005. Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biology 7:581-591.”] | |||
<br>[28] [http://pubs.acs.org/doi/pdf/10.1021/jf040466r “Christophoridou, S., P. Dais, L.H. Tseng, M. Spraul. 2005. Separation and identification of phenolic compounds in olive oil by coupling high-performance liquid chromatography with postcolumn solid-phase extraction to nuclear magnetic resonance spectroscopy (LC-SPE-NMR). Journal of Agricultural and Food Chemistry 53(12):4667-4679.”] | |||
<br>[29] [http://pubs.acs.org/doi/pdf/10.1021/jf060807w “Cayuela, J.A., M. Rada, J.J. Rios, T. Albi, A. Guinda. 2006. Changes in phenolic composition induced by Pseudomonas savastanoi pv. Savastanoi infection in olive tree: presence of large amounts of verbascoside in nodules of tuberculosis disease. Journal of Agricultural and Food Chemistry 54(15):5363-5368.”] | |||
<br>[30] [http://aem.asm.org/content/76/11/3611.full.pdf+html “Perez-Martinez, I., L. Rodriguez-Moreno, L. Lambertsen, I.M. Matas, J. Murillo, S. Tegli, A.J. Jimenez, C. Ramos. 2010. Fate of a Pseudomonas savastanoi pv. savastanoi type III secretion system mutant in olive plants (Olea europea L.). Applied and Environmental Microbiology 76(11):3611-3619.”] | |||
<br>[31] [http://www.nature.com/ismej/journal/v5/n12/pdf/ismej201165a.pdf “Hosni, T., C. Moretti, G. Devescovi, Z.R. Suarez-Moreno, M.B. Fatmi, C. Guarnaccia, S. Pongor, A. Onofri, R. Buonaurio, V. Venturi. 2011. Sharing of quorum-sensing signals and role of interspecies communities in a bacterial plant disease. Multidisciplinary Journal of Microbial Ecology 5:1857-1870.”] | |||
<br>[32] [http://www.nature.com/ismej/journal/v3/n7/pdf/ismej200930a.pdf “Dulla, G.F. and S.E. Lindow. 2009. Acyl-homoserine lactone-mediated cross talk among epiphytic bacteria modules behavior of Pseudomonas syringae on leaves. Multidisciplinary Journal of Microbial Ecology 3:825-834.”] | |||
<br>[33] [http://sipav.org/main/jpp/index.php/jpp/article/view/1223 “Krid, S., M. Bouaziz, M.A. Triki, A. Gargouri, A. Rhouma. 2011. Inhibition of olive knot disease by polyphenols extracted from olive mill waste water. Journal of Plant Pathology 93(3):561.”] | |||
<br>[34] [http://www.sciencedirect.com/science/article/pii/S0923250806002166 “Perez-Martinez, I., L. Rodriguez-Moreno, I.M. Matas, C. Ramos. 2007. Strain selection and improvement of gene transfer for genetic manipulation of Pseudomonas savastanoi isolated from olive knots. Research in Microbiology 158(1):60-69.”] | |||
<br>[35] [http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6976.2011.00269.x/pdf “Silby, M.W., C. Winstanley, S.A.C. Godfrey, S.B. Levy, R.W. Jackson. 2011. Pseudomonas genomes: diverse and adaptable. FEMS Microbiology Reviews 35(4):652-680.”] | |||
<br>[36] [http://www.sciencedirect.com/science/article/pii/S016770120200163X “Bertolini, E., R. Penyalver, A. Garcia, A. Olmos, J.M. Quesada, M. Cambra, M.M. Lopez. 2003. Highly sensitive detection of Pseudomonas savastanoi pv. savastanoi in asymptomatic olive plants by nested-PCR in a single closed tube. Journal of Microbiological Methods 52(2):261-266.”] | |||
<br>[37] [http://jb.asm.org/content/190/2/625.full.pdf+html “Perez-Martinez, I., Y. Zhao, J. Murillo, G.W. Sundin, C. Ramos. 2008. Global genomic analysis of Pseudomonas savastanoi pv. savastanoi plasmids. Journal of Bacteriology 190(2):625-635.”] | |||
<br>[38] [http://ucanr.edu/sites/oliveknot/files/86253.pdf “Fichtner, E.J. 2011. Olive Knot: Integrated Pest Management for Home Gardeners and Landscape Professionals. University of California Statewide Integrated Pest Management Program Agriculture and Natural Resources. Publication 74156.”] | |||
<br>[39] [http://www.sciencedirect.com/science/article/pii/S0261219410002346 “Quesada, J.M, R. Penyalver, J. Perez-Panades, C.I. Salcedo, E.A. Carbonell, M.M. Lopez. 2010. Comparison of chemical treatments for reducing epiphytic Pseudomonas savastanoi pv. Savastanoi populations and for improving subsequent control of olive knot disease. Crop Protection 29(12):1413-1420.”] | |||
<br>[40] [http://link.springer.com/content/pdf/10.1007%2Fs13213-011-0239-0.pdf “Krid, S., M.A. Triki, A. Gargouri, A. Rhouma. 2012. Biocontrol of olive knot disease by Bacillus subtilis isolated from olive leaves. Annals of Microbiology 62(1):149-154.”] | |||
Edited by student of [mailto:slonczewski@kenyon.edu Joan Slonczewski] for [http://biology.kenyon.edu/courses/biol238/biol238syl10.html BIOL 238 Microbiology], | Edited by student of [mailto:slonczewski@kenyon.edu Joan Slonczewski] for [http://biology.kenyon.edu/courses/biol238/biol238syl10.html BIOL 238 Microbiology], 2013, [http://www.kenyon.edu/index.xml Kenyon College]. | ||
<!--Do not edit or remove this line-->[[Category:Pages edited by students of Joan Slonczewski at Kenyon College]] | <!--Do not edit or remove this line-->[[Category:Pages edited by students of Joan Slonczewski at Kenyon College]] | ||
Latest revision as of 03:22, 10 May 2013
Hildy Joseph, Kenyon 2013
Introduction

Olive knot disease, which is characterized by knots and galls on emerging stems, branches, and occasionally leaves [Fig 1.], was first described in the fourth century B.C. by the Greek philosopher Teophrastus [Fig. 2].

Pseudomonas savastanoi is the bacteria responsible for the condition [1]. Infection occurs through wounds incurred by pruning or from severe weather events like freezing and hail [2]. A wide range of symptoms can result from infection, including death and decay. Other consequences of the pathogen include defoliation, reduced limb and overall plant size. Infections can be asymptomatic for up to eight months from exposure [1]. Knots usually form in the spring during the most active growth period [2]. Plant vulnerability is also increased in the autumn, in part because of other pathogens like the fungus Spilocaea oleaginea target olive trees concurrently. Thus, the chance of co-infection by P. savastanoi is heightened [4]. The bacterium spreads easily within an orchard via wind and rain as well as pruning tools [2]. Moreover, once infected, P. savastanoi can spread to new wound sites within the same plant [5]. Two cultivars were both inoculated with P. savastanoi to determine its spread through orchards. All trees, regardless of inoculation, were prone to infection. However, inoculated trees were significantly less vigorous over the four-year study [Fig. 3] [3].
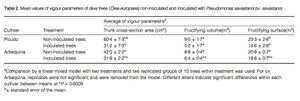
Trees that were not inoculated tended to have better health, in general [3]. Olive quality from infected plants is greatly reduced. Olives from infected plants are bitter, salty, sour, or rancid [1]. Yield is also negatively impacted by the disease. Olive knot disease is a global issue and has been found in parts of Europe, Asia, Africa, North America, South America, and Australia. The incidence and degree of virulence varies by location [2].
Understanding the virulence of P. savastanoi is important because of the emerging economic significance of olives and olive oil [Fig. 4].

Olive oil comprises a major part of the Mediterranean diet, which is becoming increasingly popular because of the associated health benefits. The Mediterranean diet may contribute to reducing LDL-cholesterol and improving HDL-cholesterol, which lessens the risk for heart disease. Digestion, autoimmune, and inflammatory diseases can all also be improved with the Mediterranean diet [6] [7] . P. savastanoi infection is part of a greater environmental concern about the high incidence of monoculture in crop orchards. Reducing genetic diversity can increase susceptibility to disease. Thus, understanding plant pathogens is essential for preserving the world's food supplies [8].
Classification of P. savastanoi
The genus Psuedomonas consists of Gram-negative bacteria that are highly prevalent in nature. Both phytopathogenic as well as nonvirulent species are part of the group, in addition to the single human pathogen, Pseudomonas aeruginosa [9] [10] [11]. P. savastonoi is a nonsporeforming bacteria that contains several flagella for motility [9]. The occurrence of phytopathogeny is polphyletic among species of Pseudomonas, which provides evidence for horizontal gene transfer of virulence genes. The virulence genes are termed the 'P. syringae complex' and may have first arisen in the common ancestor for the clade of plant pathogens within Pseudomonas, which includes P. savastanoi. More recently evolutionarily, the virulence genes may have been transferred horizontally to other species in the Pseudomonas genus. The complex comprises a pathogenicity island. [10]. Generally, pathogens in the P. syringae complex cause leaf death and cankers, and thus the olive knots caused by P. savastanoi pv. savastanoi are unique [12].
P. savastonoi was originally thought to be one of forty-five pathovars of Pseudomonas syringae due to similarities in biochemistry and physiology. DNA-DNA hybridization data revealed that Pseudomonas savastanoi actually has sufficient genetic distinction from P. syringae to merit its own species classification. Several pathovars were subsequently attributed to P. savastanoi, including P. savastanoi pv. savastanoi, P. savastanoi pv. glycinea, and P. savastanoi pv. phaseolicola [13]. Pathovars that have subsequently been recognized include P. savastanoi pv. fanxini and P. savastanoi pv. nerii.
Host range and susceptibility
Both cultivated and wild olives (Olea europaea) as well as ash (Fraxinus excelsior) are susceptible to tumorigenesis as a result of P. savastanoi pv. savastanoi. There is some overlap in hosts with other closely related pathovars. P. savastanoi pv. nerii infects oleander (Nerium oleander) but can also infect olives. P. savastanoi pv. savastanoi has been shown to infect oleander, but the occurrence is unusual [12]. Though P. savastanoi pv. savastanoi is generally limited to olive and oleander hosts, there is a recent report of a P. savastanoi infection in Mandevilla sanderi, commonly known as Brazilian jasmine [14]. There are several other pathogenic pathovars in the P. savastanoi species. P. savastanoi pv. glycinea infects soybeans and P. savastanoi pv. phaseolicola is responsible for halo blight in beans [13].
Resistant olive cultivars are rare, though the amount susceptibility to P. savastanoi does vary. Young olive plants are particularly non-resistant to infection [5]. Interestingly, P. savastanoi can be epiphytic upon infection [15].
Mechanisms of Virulence
Plant hormones: cytokinin and IAA
Infection by Pseudomonas savastanoi causes cavities in host cells in which bacteria can aggregate [Fig. 5] [16]. P. savastanoi also accumulate in the xylem, which suggests a mechanism, heretofore unverified, of bacterial transport throughout the plant [15].
Plant hormones are one aspect of virulence that activates tumorigenesis. Cytokinin and IAA production both contribute to knot formation by interfering with endogenous signals to induce rapid proliferation of host cells at the site of infection [16] [17] [18]. Pseudomonas savastanoi carry plasmids that encode for indole-3-acetic acid (IAA), a protein involved in auxin signaling [17] [19]. Various genetic studies confirm the importance of IAA and cytokinins for virulence. Mutants lacking either IAA or cytokinins were less symptomatic upon infection of a host olive plant. Without IAA or cytokinins, infections were limited to displaying only some of the normal symptoms, such as leaf necrosis and stem swelling. IAA is sufficient for tumor formation, though cytokinins also play an important role [20]. Strains lacking the pIAA1 plasmid, which encodes for the two genes responsible for IAA production, iaaM and iaaH, fail to induce olive knot growth in hosts plants [21]. Auxin may be important for virulence by downregulating factors that contribute to the host defense. For example, auxin can protect against the hypersensitive response, a defense mechanism in plants injected with P. syringae [22]. HR (hypersensitive response) is a form of programmed cell death in host cells and is widely conserved among plants and animals [23]. Auxin has many additional roles. It may regulate gene expression or affect the physiology of host cells, for example [21].
P. savastanoi occur in conjunction with other bacterial species within olive knots. It is hypothesized that phytohormones produced by the nonpathogenic bacteria coexisting in olive knots may influence P. savastanoi [12]. Auxin is produced by a variety of other genera and species that are common to olive knots, including Stenotrophomonas maltophila, Pantoea agglomerans, Pantoea oleae, Burkholderia cepacia, and Hafrnia alvei, all of which are other Gram-negative rod forming bacteria. Furthermore, IAA specifically is produced by other Pseudomonas species [24].
Suppressing host immune response and defense mechanisms
The P. Syringae complex, which is made up of the hrp/hrc gene cluster, acts on the host immune system [18] [23]. The hrp genes encode for a type III protein secretion system (T3SS) [Fig. 6]. The T3SS is comprised of a pilus that connects the bacterium with the host membrane and the pore that forms in the host cell to allow entry of type III effector proteins. All of the proteins associated with the T3SS are encoded for in the P. Syringae pathogenicity island [23]. T3SS are found in Gram-negative bacteria that interact eukaryotes, both as pathogens or symbionts, such as nitrogen-fixing Rhizobium [25]. General defense mechanisms in plants involve mechanical resistance to pathogens through the cell wall. Additionally, plants contain a set of receptors that respond to microbe-associated molecular patterns to induce production of antimicrobial agents [23]. The T3SS complex can overcome plant cell walls with the aid of helper proteins that are secreted into the extracellular space [23]. Host cell wall degradation has been observed in in olive knots [15].
The T3SS proteins evolved from a common ancestor and the genomic island encoding all of the associated proteins was transferred horizontally among seven families. P. savastanoi and other Pseudomonads exhibit the Hrc-Hrp2-T3SS family. The other genera that share T3SS-2 are Erwinia, Ralstonia, and Xanthomonas [25].
One emerging area of research is understanding the specific effectors and other elements of the T3SS [26]. The gene clusters are essential to P. savastanoi virulence and may be more important pathologically than auxin and cytokinin action. The hrp gene cluster reduces plant vitality by inducing leakage of essential nutrients into the extracellular milieu, in addition to its roles inhibiting the plant immune system. The closely related hrc genes are responsible for the plant HR [18].
In addition to the T3SS, other interactions with host defense mechanisms have been postulated. Plants produce a range of secondary metabolites that aid in defense. Flavonoids are one class of phenolic compounds that are protective against parasites and pathogens [27]. Flavonoids, along with other phenolic compounds like secoiridoids and lignans are common to olive oil and confer some of the major health benefits associated with the Mediterranean diet [7] [28]. Sequence data and empirical evidence in P. savastanoi reveals the potential for phenol catabolism. Other pathogens of woody plants, in which phenolic defense mechanisms are common, show similar catabolic abilities [26]. Furthermore, olive knots induced by P. savastanoi pv. savastanoi show a higher presence of phenolic compounds than in other leafs and shoots [29]. Thus, P. savastanoi may circumvent the the secondary metabolites produced by plants to preempt and actively combat bacterial infections by catabolizing the compounds [26].
Emerging views of virulence: signaling mechanisms and outer membrane vesicles
The P. savastanoi signaling mechanism is another quality that has been implicated in its virulence. Gram-negative bacteria commonly use the N-acyl homoserine lactone (AHL) signaling molecules. P. savastanoi often occurs in association with P. agglomerans and E. toletana, which are both nonpathogenic Enterobacteriaceae that frequently grow at olive knot infection sites [31]. In general, plants that have been inoculated with P. savastanoi have a higher overall bacteria population than plants that do not bear an infection by P. savastanoi [3]. AHL signals can be shared across P. agglomerans, E. toletana, and P. savastanoi. Specifically, the association of E. toletana and P. savastanoi confers synergistic pathogenic effects. The interactions between E. toletana and P. savastanoi are stable and lead to a higher rate of olive knot formation [31]. Alternatively, interactions between P. savastanoi and resident microbial communities on host plants may negatively impact bacterial virulence [31] [32]. The pathogenicity of P. Syringae pv. syringae, which is closely related species to P. savastanoi pathovars, was reduced by crosstalk with signaling molecules from the resident bacterial populations. Instead of synergistic effects, the AHLs from other species interfered with AHL signaling in P. Syringae [32]. Researchers are working on exploiting the interactions between resident populations and P. savastanoi to reduce its incidence and virulence [33].
Additionally, P. savastanoi employ a unique mechanism among plant pathogens by inducing the release of outer membrane vesicles from the host cell [Fig. 7]. Animal pathogens commonly release outer membrane vesicles for transporting virulence factors into host cells. The role of the membrane vesicles in a P. savastanoi infection may be a relating to correcting for osmotic pressure [30].
Development of Genetic Tools for Understanding Virulence
Major advances in the last several years have rapidly progressed the understanding of P. savastanoi after decades of stalled research. Before 2007, the significant impediment to studying P. savastanoi was the lack of any strains that were amenable to genetic manipulation, specifically conjugation. The problem is consistent across other Pseudomonads that infect woody plants, including P. savastanoi pvs. Savastanoi, franxini, and nerii. Several strains were isolated that are have more efficient transformations [34]. One of the strains with the highest transformation efficiency, P. savastanoi pv. savastanoi NCPPB 3335, was recently sequenced [26] [34]. A draft genome was obtained with pyrosequencing and compared to other published P. Syringae genomes. The primary goal of the genetic manipulation is to augment the current poor understanding of P. savastanoi virulence [26].
The draft genome of P. savastanoi pv. savastanoi NCPPB 3335 revealed 73 genes that are not found in other species as well as eight gene inversions in comparison with closely related species. Researchers suggest that the basis for host specificity is encoded for in the 73 genes unique to P. savastanoi [26] [35].
Other emerging techniques have recently allowed for easier plant manipulation and greater resolution. For example, cultivation by micropropagation circumvents the arduous growing requirements for olive trees, including long growing seasons and large plant size [Fig. 8]. Additionally, the draft P. savastanoi pv. savastanoi NCPPB 3335 genome has been employed to develop a green fluorescent protein tagging system. Scanning and transmission electron microscopy can be used to visualize the bacteria in planta and further elucidate virulence mechanisms [12].

Responding to Olive Knot Disease in Crop Management
Detection of P. Savastanoi
An infection can easily be recognized on crop plants in its late stages. However, insect wounds are easily confused with olive knots in the preliminary onset. Bacteria can be isolated from knots and cultured in the laboratory for identification and confirmation, though older knots generally contain a variety of secondary invaders [2]. Moreover, plants can be infected without having visible symptoms [36]. Furthermore, P. savastanoi grows relatively slowly in the laboratory, with a required incubation between two to three days at 27°C [2] [9]. Other techniques have been developed to rapidly detect P. savastanoi without culturing or the necessity of serological methods. For example, there is a highly sensitive PCR technique based on amplification of the iaaL gene, which encodes for a product that catalyzes the addition of lysine to IAA [36]. Isolates can be distinguished by sequence differences, specifically differing numbers of nucleotide motif tandem repeats, in both iaaL paralogs found in different P. savastanoi strains [37]. Another technique using rt-PCR can simultaneously detect P. savastanoi along with four RNA viruses from a single host sample. Detecting multiple plant pathogens in a single reaction conserves time and resources [6].
Control of the pathogen
Farmers have a variety of options to alleviate the problem of olive knot disease, though noncommercial farmers have fewer available products. Landscapers and homeowners are advised to practice several strategic planting practices to minimize the chances for disease dissemination. For example, timing of olive tree purchase is critical, as P. savastanoi is often epiphytic during the winter months and infections are most visible in late spring. Furthermore, visible galls should be removed with sterile pruning tools to minimize spread of the bacteria [38]. The levels of epiphytic bacteria may be positively correlated with the occurrence of olive knots, and thus minimizing the presence of olive knots reduces the overall spread of the infection [39]. One measure to preemptively combat disease is to remove oleander plants that are growing in close proximity to growing area because of the potential overlap in host specificity for P. savastanoi [38]. However, P. savastanoi is generally an obligate symbiont and does not proliferate on soil [12].
Resistant olive cultivars would be the best method for controlling P. savastanoi. However, naturally resistant cultivars have not yet been detected. Recently, susceptibility measures to P. savastanoi have been standardized, which will aid in developing cultivars that will not become infected [12].
Chemical
Copper compounds are commonly used to reduce P. savastanoi proliferation [33]. The Cu2+ ions can be toxic to P. savastanoi. Copper can also induce a viable but nonculturable state, which interferes with serological detection of P. savastanoi on host plants [39]. The method poses threats to the environment and public health. Furthermore, control of P. savastanoi with copper shows reduced efficacy with continued use [33]. A couple copper compounds are available, including copper oxychloride and cuprocalcic suflate plus mancozeb, which have been shown to be equally effective in reducing the amount of P. savastanoi growing on the leafs and stems [39]. Maximal efficacy of treatment requires a minimum of biannual application in the spring and fall, when olive trees are at the highest risk for infection. Olive trees are predisposed for infection when leafs fall in the spring due to residual leaf scarring as well as during frosts in the late autumn [4]. Bacteria that resist the copper treatment and exist in epiphytic and endophytic states can proliferate with failure to adhere to a regular copper treatment schedule [39]. However, farmers are recalcitrant toward biannual copper application because of its effect on the olive product [4].
Polyphenols have also demonstrated efficacy in controlling olive knot. The compounds were obtained in extracts of olive mill waste water. Even at the lowest concentration tested, 100 mg/L, all olive knot formation was blocked, whereas a concurrent copper treatment was less effective [33].
Biological
Other bacterial species can be used to control the incidence of P. savastanoi. Antimicrobial proteins produced by other microbes include colicin-like bacteriocins. Bacteriocins are common bacterial protein products that are detrimental to closely related species. Bacteriocins from P. Syringae pv. ciccaronei can inhibit both olive knot growth as well as epiphytic P. savastoni infection [8]. Another promising development in the biological control of olive blot is the application of Bacillus subtilis. B. subtilis produces antibiotics like zwittermicin-A and kanosamine as well as other antimicrobial lipoproteins. A protein produced from B. subtilis has been shown to reduce the size of olive knots caused by P. savastanoi [40].
Conclusion
Olive knot disease is of great economic importance due to the increasing interest in the benefits of polyunsaturated fatty acids in the diet. Farmers maximize profit and minimize the necessity of environmentally harmful treatments if pests can be controlled. P. savastanoi is the major threat to olive trees. Exciting breakthroughs have occurred in recent years relating to the genetic manipulation of P. savastanoi, which will allow for a greater understanding of the plant pathogen's virulence.
References
[1] Iacobellis NS. 2001. Olive knot. In: Maloy OC, Murray TD, eds. Encyclopedia of plant pathology. New York, NY, USA: John Wiley & Sons, 7143–715.
[2] "Young, J.M. 2004. Olive knot and its pathogens. Australian Plant Pathology 33:33-39."
[3] "Quesada, J.M., R. Penyalver, J. Perez-Panades, C.I. Salcedo, E.A. Carbonell, M.M. Lopez. 2009. Dissemination in Pseudomonas savastanoi pv. savastanoi populations and subsequent appearance of olive knot disease. Plant Pathology 59(2):262-269."
[4] "Teviotdale, B.L., W.H. Krueger. 2004. Effects of copper sprays, defoliation, rainfall, and inoculum concentration on incidence of olive knot disease. Plant Disease 88:131-135."
[5] "Penyalver, R., A. Garcia, A. Ferrer, E. Bertolini, J.M. Quesada, C.I. Salcedo, J. Piquer, J. Perez-Panades, E.A. Carbonell, C. del Rio, J.M. Caballero, M.M. Lopez. 2006. Factors affecting Pseudomonas savastanoi pv. savastanoi plant inoculaitons and their use for evaluation of olive cultivar susceptibility. Phytopathology 96(3):313-319."
[6] "Bertolini, E., A. Olmos, M.M. Lopez, M. Cambra. 2003. Multiplex nested reverse transcription-polymerase chain reaction in a single tube for sensitive and simultaneous detection of four RNA viruses and Psuedomonas savastanoi pv. savastanoi in olive trees. Phytopathology 93(3):286-292."
[7] "Alarcon de la Lastra, C., M.D. Barranco, V. Motilva, J.M. Herrerias. 2001. Mediterranean diet and health: biological importance of olive oil. Current Pharmaceutical Design 7(10):933-950."
[8] "Grinter, R., J. Milner, D. Walker. 2012. Bacteriocins active against plant pathogenic bacteria. Biochemical Society Transactions 40(6):1498-1502."
[9] "Janse, J.D. 1982. Pseudomonas syringae subsp. Savastanoi (ex Smith) subsp. Nov., nom. Rev., the bacterium casuing excrescences on Oleaceae and Merium oleander L. International Journal of Systemic and Evolutionary Microbiology 32(2):166-169."
[10] "Yamamoto, S., H. Kasai, D.L. Arnold, R.W. Jackson, A. Vivian, S. Harayama. 2000. Phylogeny of the genus Psuedomonas: intrageneric strucure reconstructed from the nucleotide sequences of gryB and rpoD genes."
[11] "Hancock, R.E.W. 1998. Resistance mechansims in Psuedomonas aeruginosa and other nonfermentative gram-negative bacteria. Clinical Infectious Diseases 27(S1):S93-S99."
[12] "Ramos, C., I.M Matas, L. Bardaji, I.M. Argon, J. Murillo. 2012 Pseudomonas savastanoi pv. savastanoi: some like it knot. Molecular Plant Pathology 13(9):998-1009."
[13] "Gardan, L., C. Bollet, M. Abu Ghorrah, F. Grimont, P.A.D. Grimont. 1992. DNA relatedness among the pathovar strains of Pseudomonas syringae subsp. Savastanoi Janse (1982) and proposal of Psuedomonas savastanoi sp. nov. International Journal of Systemic and Evolutionary Microbiology 42(4):606-612."
[14] “Eltlbany, N., Z.Z. Prokscha, M.P. Castaneda-Ojeda, E. Krogerrecklenfort, H. Heuer, W. Wohanka, C. Ramos, K. Smalla. 2012. A new bacterial disease on Mandevilla sanderi, caused by Pseudmonas savastanoi: lessons learned for bacterial diversity studies. Applied Environmental Microbiology 78(23):8492-8297.”
[15] “Rodriguez-Moreno, L., A.J. Jimenez, C. Ramos. 2009. Endopathogenic lifestyle of Pseudomonas savastanoi pv. Savastanoi in olive knots. Microbial Biotechnology 2(4):476-488.”
[16] “Marchi, G., B. Mori, P. Pollacci, M. Mencuccini, G. Surico. 2009. Systemic spread of Pseudomonas savastanoi pv. Savastanoi in olive explants. Plant Pathology 58:152-158.”
[17] “Surico, G., N.S. Iacobellis, A. Sisto. 1985. Studies on the role of indole-3-acetic acid and cytokinins in the formation of knots on olive and oleander plants by Pseudomonas syringae pv. Savastanoi. Physiological Plant Pathology 26:309-320.”
[18] “Sisto, A., M.G. Cipriani, M. Morea. 2004. Knot formation caused by Pseudomonas syringae subsp. savastanoi on olives is hrp-dependent. Phytopathology 94(5):484-489.”
[19] “Muto, H., M.K. Watahiki, K.T. Yamamoto. 2007. What makes each Aux/IAA gene unique in its gene family, expression pattern or properties of the gene product? Plant Signaling and Behavior. 2(5):390-392.”
[20] “Iacobellis, N.S., A. Sisto, G. Sucrico, A. Evidente, E. DiMaio. 1994. Pathogenicity of Pseudomonas syringae subsp. savastanoi mutants defective in phytohormone production. Journal of Phytopathology 140(3):238-248.”
[21] “Spaepen, S., J. Vanderleyden. 2010. Auxin and plant-microbe interactions. Cold Spring Harbor Perspectives in Biology 3(4):a001438.”
[22] “Melotto, M., B.N. Kunkel. 2013. Virulence strategies of plant pathogenic bacteria. In: The Prokaryotes – Prokaryotic Physiology and Biochemistry, eds. E. Rosenberg et al. Springer-Verlag, Berlin, pp. 61-82.”
[23] “Grant, S.R., E.J. Fisher, J.H. Chang, B.M. Mole, J.L. Dangl. 2006. Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annual Reviews of Microbiology 60:425-449.”
[24] “Ouzari, H., A. Khsairi, N. Raddadi, L. Jaoua, A. Hassen, M. Zarrouck, D. Daffonchio, A. Boudabous. 2008. Diversity of auxin-producing bacteria associated to Pseudomonas savastanoi-induced olive knots. Journal of Basic Microbiology 48:370-377.”
[25] “Gazi, A.D., P.F. Sarris, V.E. Fadouloglou, S.N. Charova, N. Mathioudakis, N.J. Panopoulos, M. Kokkinidis. 2012. Phylogenetic analysis of a gene cluster encoding an additional, rhizobial-like type III secretion system that is narrowly distributed among Pseudomonas syringae strains. BMC Microbiology 12:188.”
[26] “Rodriguez-Palenzuela, P., I.M. Matas, J. Murillo, E. Lopez-Solanilla, L. Bardaji, I. Perez-Martinez, M.E. Rodriguez-Moskera, R. Penyalver, M.M. Lopez, J.M. 2010. Annotation and overview of the Pseudomonas savastanoi pv. savastanoi NCPPB 3335 draft genome reveals the virulence gene complement of a tumour-inducing pathogen of woody hosts. Environmental Microbiology 12(6):1604-1620.”
[27] “Treutter, D. 2005. Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biology 7:581-591.”
[28] “Christophoridou, S., P. Dais, L.H. Tseng, M. Spraul. 2005. Separation and identification of phenolic compounds in olive oil by coupling high-performance liquid chromatography with postcolumn solid-phase extraction to nuclear magnetic resonance spectroscopy (LC-SPE-NMR). Journal of Agricultural and Food Chemistry 53(12):4667-4679.”
[29] “Cayuela, J.A., M. Rada, J.J. Rios, T. Albi, A. Guinda. 2006. Changes in phenolic composition induced by Pseudomonas savastanoi pv. Savastanoi infection in olive tree: presence of large amounts of verbascoside in nodules of tuberculosis disease. Journal of Agricultural and Food Chemistry 54(15):5363-5368.”
[30] “Perez-Martinez, I., L. Rodriguez-Moreno, L. Lambertsen, I.M. Matas, J. Murillo, S. Tegli, A.J. Jimenez, C. Ramos. 2010. Fate of a Pseudomonas savastanoi pv. savastanoi type III secretion system mutant in olive plants (Olea europea L.). Applied and Environmental Microbiology 76(11):3611-3619.”
[31] “Hosni, T., C. Moretti, G. Devescovi, Z.R. Suarez-Moreno, M.B. Fatmi, C. Guarnaccia, S. Pongor, A. Onofri, R. Buonaurio, V. Venturi. 2011. Sharing of quorum-sensing signals and role of interspecies communities in a bacterial plant disease. Multidisciplinary Journal of Microbial Ecology 5:1857-1870.”
[32] “Dulla, G.F. and S.E. Lindow. 2009. Acyl-homoserine lactone-mediated cross talk among epiphytic bacteria modules behavior of Pseudomonas syringae on leaves. Multidisciplinary Journal of Microbial Ecology 3:825-834.”
[33] “Krid, S., M. Bouaziz, M.A. Triki, A. Gargouri, A. Rhouma. 2011. Inhibition of olive knot disease by polyphenols extracted from olive mill waste water. Journal of Plant Pathology 93(3):561.”
[34] “Perez-Martinez, I., L. Rodriguez-Moreno, I.M. Matas, C. Ramos. 2007. Strain selection and improvement of gene transfer for genetic manipulation of Pseudomonas savastanoi isolated from olive knots. Research in Microbiology 158(1):60-69.”
[35] “Silby, M.W., C. Winstanley, S.A.C. Godfrey, S.B. Levy, R.W. Jackson. 2011. Pseudomonas genomes: diverse and adaptable. FEMS Microbiology Reviews 35(4):652-680.”
[36] “Bertolini, E., R. Penyalver, A. Garcia, A. Olmos, J.M. Quesada, M. Cambra, M.M. Lopez. 2003. Highly sensitive detection of Pseudomonas savastanoi pv. savastanoi in asymptomatic olive plants by nested-PCR in a single closed tube. Journal of Microbiological Methods 52(2):261-266.”
[37] “Perez-Martinez, I., Y. Zhao, J. Murillo, G.W. Sundin, C. Ramos. 2008. Global genomic analysis of Pseudomonas savastanoi pv. savastanoi plasmids. Journal of Bacteriology 190(2):625-635.”
[38] “Fichtner, E.J. 2011. Olive Knot: Integrated Pest Management for Home Gardeners and Landscape Professionals. University of California Statewide Integrated Pest Management Program Agriculture and Natural Resources. Publication 74156.”
[39] “Quesada, J.M, R. Penyalver, J. Perez-Panades, C.I. Salcedo, E.A. Carbonell, M.M. Lopez. 2010. Comparison of chemical treatments for reducing epiphytic Pseudomonas savastanoi pv. Savastanoi populations and for improving subsequent control of olive knot disease. Crop Protection 29(12):1413-1420.”
[40] “Krid, S., M.A. Triki, A. Gargouri, A. Rhouma. 2012. Biocontrol of olive knot disease by Bacillus subtilis isolated from olive leaves. Annals of Microbiology 62(1):149-154.”
Edited by student of Joan Slonczewski for BIOL 238 Microbiology, 2013, Kenyon College.