Chromohalobacter Salexigens: Difference between revisions
Sydneyrodman (talk | contribs) No edit summary |
|||
| (40 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | |||
{{Biorealm Genus}} | {{Biorealm Genus}} | ||
| Line 6: | Line 7: | ||
Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Halomonadaceae; Chromohalobacter; | Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Halomonadaceae; Chromohalobacter; | ||
[[File:Sigs.2285059-f2.jpg|thumb|right|400px|"Chromohalobacter salexigens"[[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3368415/figure/f2/]]]] | |||
{| | {| | ||
| Line 14: | Line 18: | ||
===Species=== | ===Species=== | ||
C. | ''C. salexigens'' | ||
==Description and significance== | ==Description and significance== | ||
This bacterium is a moderate halophile, | "C. salexigens" is a gram negative marine bacterium that lives in hypersaline environments. It was isolated from the island of Bonaire, Netherlands Antilles [6].This bacterium is a moderate halophile, with a broad salinity range. ''C. salexigens'' is very flexible in that its salt requirements can be met by ions of other salts such as potassium, rubidium, ammonium, bromide, and others [2]. It is an aerobic chemoorganotroph. It can live in a variety of saline conditions but their optimum concentration is between 2-2.5M [6]. | ||
==Genome structure== | ==Genome structure== | ||
DNA Bases: 3696649 | DNA Bases: 3696649 | ||
| Line 39: | Line 37: | ||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
The | The ability of ''C. salexigens'' to survive in a broad range of salinity, makes it a euryhaline bacterium [6]. "C. salexigens" can be considered extremophiles because they are able to survive in hypersaline conditions due to its osmoregulatory mechanisms, such as its production of ectoine. Its versatile metabolism allows for fast growth because of its ability to use a variety of simple carbon compounds as both its carbon and energy source. This bacterium compared, to other organisms that can live in high saline environments, has a more acidic amino acids that enable it to produce the organic solute, ectoine [7]. | ||
==Ecology== | ==Ecology== | ||
Interactions between ''C. salexigens'' and other bacterium such as various strands of Salmonella allow for salinity tolerance modulation. In other words, this bacterium allows for other organisms to exist in environments they would otherwise not be able to cope with [4]. | |||
"C. salexigens" have the eight- gene cluster in order to from Isethionate from taurine, one of the simple compounds that this bacterium uses for its carbon and energy source. The formation of isethionate is used as nutrients for bacterium, "C. salexigens" create a reserve to use as nutrients for other bacterium [8]. | |||
==Pathology== | ==Pathology== | ||
C. | Current research indicates that ''C. salexigens'' is not known to be pathogenic. | ||
==Application to Biotechnology== | ==Application to Biotechnology== | ||
In response to salt and temperature stress ''C. salexigens'' produces and stores various solutes. Some solutes, namely hydroxyectoine, are used in thermoregulation processes that protect ''C. salexigens'' from extreme temperatures. [3] | |||
==Current Research== | ==Current Research== | ||
N(gamma)-acetyl-2,4-diaminobutyrate (NADA), | [[File:3034673_1752-0509-5-12-5.png |thumb|right|400px|Ectoine biosynthesis pathways[[http://openi.nlm.nih.gov/detailedresult.php?img=3034673_1752-0509-5-12-5&query=the&fields=all&favor=none&it=none&sub=none&uniq=0&sp=none&req=4&simCollection=2795831_11999_2009_881_Fig5_HTML&npos=2&prt=3]]]] | ||
"C. salexigens" are used as the model organism to test osmoadaptation, ectoine is a natural compound found in this bacterium and "C. salexigens" are osmoadaptive because of its' ability to biosynthesize ectoine [7]. | |||
Scientists are researching mutant ''C. salexigens'' bacterium that synthesize ectoine. ''C. Salexigens'' mutants can be used to produce N(gamma)-acetyl-2,4-diaminobutyrate (NADA). When mutants are placed in conjunction with the bacterium ''Salmonella Enterica Serovar Typhimurium'', salinity stress typically present in this form of Salmonella ceased to persist. [4] | |||
In other research, ''C. Salexigens'' is being investigated to better understand its long-term response to salinity stress regarding membrane modulation. Ectoine-deficient strains of ''C. salexigens'' are unable to cope with salinity stress and undergo extensive membrane changes. The addition of ectoine to these deficient strains, however, allows these bacterium to maintain a salinity responsive membrane. [5] | |||
==References== | ==References== | ||
[http://wishart.biology.ualberta.ca/BacMap/index.html Stothard P, Van Domselaar G, Shrivastava S, | [1] [http://wishart.biology.ualberta.ca/BacMap/index.html Stothard P, Van Domselaar G, Shrivastava S, et. al. 2005. BacMap: an interactive picture atlas of annotated bacterial genomes. Nucleic Acids Res 33:D317-D320] | ||
[2] [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=14532102&query_hl=2&itool=pubmed_docsum O'Connor K, Csonka LN. 2003. Salt Requirements of C. Salexigens. Department of Biological Sciences, Purdue University, West Lafayette, Indiana 47907-1392.] | |||
[3] [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=16707670&query_hl=2&itool=pubmed_docsum Garcia-Estepa R, Argandona M, Reina-Bueno M, et. al. 2006. Thermoprotection of C. Salexigens. Department of Microbiology and Parasitology, Faculty of Pharmacy, University of Seville, Spain.] | |||
[4] [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=16469465&query_hl=2&itool=pubmed_docsumGarcia-Estepa Garcia-Estepa R, Canovas D, Iglesias-Guerra F, et. al. 2006. Osmoprotection of Salmonella Enterica. Department of Microbiology and Parasitology, Faculty of Pharmacy, University of Seville, Spain.] | |||
[5] [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=16156114&query_hl=2&itool=pubmed_docsumGarcia-Estepa Vargas C, Kallimanis A, Koukkou AI, et. al. 2005. NADA, the Precursor to Ectoine. Department of Microbiology and Parasitology, University of Seville, Spain.] | |||
[6] [http://www.springerlink.com/content/v2tmu33122208324/ Oren, A, Larimer F, Richardson P, et al. 2005. How to be moderately halophilic with broad salt tolerance. Extremophiles. 9:275-279.] | |||
[7] [http://www.biomedcentral.com/1752-0509/5/12 Ates O, Toksoy E, Arga K. 2011. Genome-scale reconstruction of metabolic network for a halophilic extremophile Chromohalobacter salexigens DSM 3043. BMC Systems Biology 5:12.] | |||
[http:// | [8] [http://mic.sgmjournals.org/content/156/5/1547.full Krejcik Z, Hollemeyer K, Smits T, Cook A. 2010. Isethionate formation from taurine in CHromohalobacter salexigens: purification of sulfoacetaldehyde reductase. Microbiology 156: 1547-1555.] | ||
Edited by Chris Wittrock, a student of [mailto:ralarsen@ucsd.edu Rachel Larsen] and Kit Pogliano | Edited by Chris Wittrock, a student of [mailto:ralarsen@ucsd.edu Rachel Larsen] and Kit Pogliano | ||
Revised by Sydney Rodman of Randolph-Macon College | |||
Latest revision as of 00:17, 5 December 2012
A Microbial Biorealm page on the genus Chromohalobacter Salexigens
Classification
Higher order taxa
Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Halomonadaceae; Chromohalobacter;
Species
C. salexigens
Description and significance
"C. salexigens" is a gram negative marine bacterium that lives in hypersaline environments. It was isolated from the island of Bonaire, Netherlands Antilles [6].This bacterium is a moderate halophile, with a broad salinity range. C. salexigens is very flexible in that its salt requirements can be met by ions of other salts such as potassium, rubidium, ammonium, bromide, and others [2]. It is an aerobic chemoorganotroph. It can live in a variety of saline conditions but their optimum concentration is between 2-2.5M [6].
Genome structure
DNA Bases: 3696649
Chromosome Type: Circular
Total Genes: 3403
Protein Coding Genes: 3319 RNA Genes: 84 Pseudo Genes: 21
Cell structure and metabolism
The ability of C. salexigens to survive in a broad range of salinity, makes it a euryhaline bacterium [6]. "C. salexigens" can be considered extremophiles because they are able to survive in hypersaline conditions due to its osmoregulatory mechanisms, such as its production of ectoine. Its versatile metabolism allows for fast growth because of its ability to use a variety of simple carbon compounds as both its carbon and energy source. This bacterium compared, to other organisms that can live in high saline environments, has a more acidic amino acids that enable it to produce the organic solute, ectoine [7].
Ecology
Interactions between C. salexigens and other bacterium such as various strands of Salmonella allow for salinity tolerance modulation. In other words, this bacterium allows for other organisms to exist in environments they would otherwise not be able to cope with [4].
"C. salexigens" have the eight- gene cluster in order to from Isethionate from taurine, one of the simple compounds that this bacterium uses for its carbon and energy source. The formation of isethionate is used as nutrients for bacterium, "C. salexigens" create a reserve to use as nutrients for other bacterium [8].
Pathology
Current research indicates that C. salexigens is not known to be pathogenic.
Application to Biotechnology
In response to salt and temperature stress C. salexigens produces and stores various solutes. Some solutes, namely hydroxyectoine, are used in thermoregulation processes that protect C. salexigens from extreme temperatures. [3]
Current Research
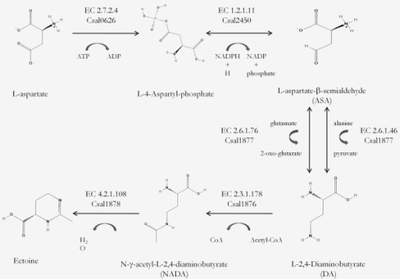
"C. salexigens" are used as the model organism to test osmoadaptation, ectoine is a natural compound found in this bacterium and "C. salexigens" are osmoadaptive because of its' ability to biosynthesize ectoine [7].
Scientists are researching mutant C. salexigens bacterium that synthesize ectoine. C. Salexigens mutants can be used to produce N(gamma)-acetyl-2,4-diaminobutyrate (NADA). When mutants are placed in conjunction with the bacterium Salmonella Enterica Serovar Typhimurium, salinity stress typically present in this form of Salmonella ceased to persist. [4]
In other research, C. Salexigens is being investigated to better understand its long-term response to salinity stress regarding membrane modulation. Ectoine-deficient strains of C. salexigens are unable to cope with salinity stress and undergo extensive membrane changes. The addition of ectoine to these deficient strains, however, allows these bacterium to maintain a salinity responsive membrane. [5]
References
Edited by Chris Wittrock, a student of Rachel Larsen and Kit Pogliano
Revised by Sydney Rodman of Randolph-Macon College
