Medical Bioremediation: Difference between revisions
| (2 intermediate revisions by the same user not shown) | |||
| Line 72: | Line 72: | ||
[[Image:7KC.jpeg|thumb|150px|left|Molecular structure of the pathogenic oxysterol 7-Ketocholesterol. <sup>[http://www.ncbi.nlm.nih.gov/pubmed/22447444 38]</sup>]] | [[Image:7KC.jpeg|thumb|150px|left|Molecular structure of the pathogenic oxysterol 7-Ketocholesterol. <sup>[http://www.ncbi.nlm.nih.gov/pubmed/22447444 38]</sup>]] | ||
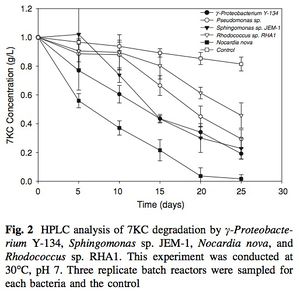
[[Image:HPLC_7KC.jpg|thumb|300px|right| HPLC Analysis of 7KC catabolysis rates in various microbes. <i>Nocardia nova</i> was the fastest degrader.<sup>[http://www.ncbi.nlm.nih.gov/pubmed/22447444 38]</sup>]] | [[Image:HPLC_7KC.jpg|thumb|300px|right| HPLC Analysis of 7KC catabolysis rates in various microbes. <i>Nocardia nova</i> was the fastest degrader.<sup>[http://www.ncbi.nlm.nih.gov/pubmed/22447444 38]</sup>]] | ||
7-ketocholesterol (7KC) is an oxidized derivative of cholesterol, a plasma membrane stabilizer and hormone precursor. 7KC is suspected of involvement in both atherosclerosis,<sup>26</sup> macular degeneration <sup>27</sup>and Alzheimer’s <sup>28</sup> as a pro-oxidant and pro-inflammatory. <sup>29</sup> 7KC is the most commonly consumed dietary [http://www.ncbi.nlm.nih.gov/pubmed/10617772 oxysterol] and has been shown to accumulate in human plasma. <sup>30</sup> Mammalian enzymes are capable of degrading 7KC, but they’re not found in the lysosome. | 7-ketocholesterol (7KC) is an oxidized derivative of cholesterol, a plasma membrane stabilizer and hormone precursor. 7KC is suspected of involvement in both atherosclerosis,<sup>26</sup> macular degeneration <sup>27</sup>and Alzheimer’s <sup>28</sup> as a pro-oxidant and pro-inflammatory agent. <sup>29</sup> 7KC is the most commonly consumed dietary [http://www.ncbi.nlm.nih.gov/pubmed/10617772 oxysterol] and has been shown to accumulate in human plasma. <sup>30</sup> Mammalian enzymes are capable of degrading 7KC, but they’re not found in the lysosome. | ||
Bacteria do not have cholesterol, they | Bacteria do not have cholesterol, they have structurally similar “surrogate” compounds called hopanoids, which confer membrane stability in a similar fashion. <sup>31</sup> Despite this absence, Mathieu et al. (2008) demonstrated that several microbial species are capable of degrading cholesterol as a sole carbon source, including <i>γ-Proteobacterium Y-134, Sphingomonas sp. JEM-1, Nocardia nova, Rhodococcus sp. RHA1</i> and <i>Pseduomonas aeruginosa. </i> <sup>32, 33</sup> <i>Nocardia nova</i> was most effective, degrading nearly 100% of the 7KC substrate in 25 days (30°C, pH 7). The runners up, <i>γ – Proteobacterium</i> and <i>Sphingomonas</i> were only able to catabolize about 80% at 25 days. This discrepancy may be due to <i>Nocardia’s</i> ability to produce [http://www.iisc.ernet.in/currsci/jul10/articles19.htm biosurfactants] <sup>34</sup> that dissolve the hydrophobic 7KC and increase the surface area available for enzymatic binding. | ||
These microbes possess oxygenase enzymes that bind molecular oxygen with hydrogen and attach the hydroxyl groups onto highly reduced aromatics. As these new hydroxyl groups are attached, the molecule becomes susceptible to dehydrogenase and hydroxylase attacks. <sup>35</sup> This may tip the scale and enable mammalian enzymes to act on the sterol substrate. | These microbes possess oxygenase enzymes that bind molecular oxygen with hydrogen and attach the hydroxyl groups onto highly reduced aromatics. As these new hydroxyl groups are attached, the molecule becomes susceptible to dehydrogenase and hydroxylase attacks. <sup>35</sup> This may tip the scale and enable mammalian enzymes to act on the sterol substrate. | ||
| Line 94: | Line 94: | ||
====Catabolysis of Macular Degeneration-related compound A2E==== | ====Catabolysis of Macular Degeneration-related compound A2E==== | ||
A2E is a major constituent of pigment lipofuscin, which builds in retinal epithelium cells with age. Dr. John Schloendorn and the team of Dr. Janet Sparrow collaborated to show that certain enzymes were capable of catabolizing the particularly sturdy A2E. These include: Crocus sativus zeaxanthin cleavage dioxygenase, cyanobacterial carotenoid cleavage dioxygenase NSC1, horseradish peroxidase (HRP) and several peroxidases. The researchers further showed that HRP could be targeted to human lysosomes in culture and there could non-toxically degrade A2E. <sup>57</sup> | A2E is a major constituent of pigment lipofuscin, which builds in retinal epithelium cells with age. Dr. John Schloendorn and the team of Dr. Janet Sparrow collaborated to show that certain enzymes were capable of catabolizing the particularly sturdy A2E. These include: Crocus sativus zeaxanthin cleavage dioxygenase, cyanobacterial carotenoid cleavage dioxygenase NSC1, horseradish peroxidase (HRP) and several peroxidases. The researchers further showed that HRP could be targeted to human lysosomes in culture and there could non-toxically degrade A2E. <sup>56, 57</sup> | ||
[[Image:HRP1.jpg|thumb|450px|center| Effect of pH on horseradish peroxidase degradation of bisretinoid A2E. Lysosomes have a pH of about 5.2, meaning HRP may be suited to the acidity. Courtesy JACS (2010). <sup>[http://www.ncbi.nlm.nih.gov/pubmed/21166406 | [[Image:HRP1.jpg|thumb|450px|center| Effect of pH on horseradish peroxidase degradation of bisretinoid A2E. Lysosomes have a pH of about 5.2, meaning HRP may be suited to the acidity. Courtesy JACS (2010). <sup>[http://www.ncbi.nlm.nih.gov/pubmed/21166406 57]</sup>]] | ||
====Amyloid Beta Degradation by <i>Mycoplasma hyorhinis</i>==== | ====Amyloid Beta Degradation by <i>Mycoplasma hyorhinis</i>==== | ||
| Line 158: | Line 158: | ||
4. Leader, B. et al. (2008) [http://www.ncbi.nlm.nih.gov/pubmed/18097458 Protein therapeutics: a summary and pharmacological classification.] Nat. Rev. Drug Discov. 7(1): 21–39 | 4. Leader, B. et al. (2008) [http://www.ncbi.nlm.nih.gov/pubmed/18097458 Protein therapeutics: a summary and pharmacological classification.] Nat. Rev. Drug Discov. 7(1): 21–39 | ||
5. | 5. Gale, E.F. (1952). [http://archive.org/details/chemicalactiviti00gale The Chemical Activities of Bacteria.] New York, Academic Press. | ||
6. de Grey, A. et al. (2005) [http://www.ncbi.nlm.nih.gov/pubmed/16040282 Medical bioremediation: Prospects for the application of microbial catabolic diversity to aging and several major age-related diseases.] Ageing Res Rev. 4(3):315-38. | 6. de Grey, A. et al. (2005) [http://www.ncbi.nlm.nih.gov/pubmed/16040282 Medical bioremediation: Prospects for the application of microbial catabolic diversity to aging and several major age-related diseases.] Ageing Res Rev. 4(3):315-38. | ||
Latest revision as of 21:57, 24 August 2013
Medical bioremediation is the technique of applying microbial xenoenzymes in human therapy. The process involves screening for enzymes capable of catabolizing the target pathogenic substrate, engineering microbes to express sufficient quantities of the enzyme and finally delivering the enzyme to the appropriate tissue and cell types.
Introduction
Bioremediation, Microbial Synthesis and Gale's Infallibility Hypothesis
Bioremediation is the technique of using organisms to catabolize toxic waste such as oil spills or industrial runoff. The most commonly used organisms are microbes, though phytoremediation is also used. 1 Wild-type microbes have proven capable of digesting highly toxic and stable compounds, but organisms can be genetically engineered to augment their ability. For example, Deinococcus radiodurans, the most radio-resistant organism known, has been modified to digest toluene and ionic mercury. 2
Microbes are the source of approximately 22,500 bioactive drug compounds. Of these, 17% were from unicellular bacteria (mainly Pseudomonas and Bacillus), 45% from filamentous bacteria (actinomycetes) and 38% from fungi. 3 Microbes are the predominant source of manufactured protein, ever since microbial human insulin production began 25 years ago. There are presently more than 130 protein therapeutics used worldwide and many more undergoing clinical trials. 4
Organic, energy-rich molecules introduced to the environment are potential microbial nutrients. The “microbial infallibility hypothesis,” coined by microbiologist Dr. Ernest Gale in 1952, 5 states that the buildup of compounds initially resistant to biodegradation exerts a strong selective pressure on nearby microbes to evolve to consume them.
Strategies for Engineered Negligible Senescence (SENS)
In 2002, Cambridge biogerontologist Dr. Aubrey de Grey theorized that the principles of bioremediation and Gale’s hypothesis could be applied to human pathology as a part of his seven-part longevity protocol, Strategies for Engineered Negligible Senescence (SENS), which aims to address: 6
1. Cell loss or atrophy (without replacement) 7
2. Oncogenic nuclear mutations and epimutations 8
3. Cell senescence (death-resistant cells) 9
4. Mitochondrial mutations 10
5. Intracellular junk (particularly lysosomal aggregates) 11
6. Extracellular aggregates 9
7. Random extracellular cross-linking (Advanced glycation end products) 12
Medical bioremediation is applicable to multiple protocols but particularly number 5, termed "LysoSENS." Recognizing that bone is all that remains of the deceased in graveyards, de Grey made it obvious that there must be soil microbes capable of catabolizing all of the organic compounds comprising the human body. Certain such compounds are pathogenic, including amyloids, neurofibriliary tangles, cholesterol, lipofuscin, huntingtin, alpha-synuclein and many others. The rapid selective pressure on microbes in the presence of these pathogenic compounds as food sources may yield therapeutic new enzymes. De Grey coined the term “medical bioremediation” to describe this emerging therapy.
The Machinery of Cellular Catabolism
Proper lysosomal functioning is paramount to the integrity of the cell, and with it longevity. Cellular components must constantly be recycled due to damage and poor construction. Proteasomes are responsible for the simpler catabolic jobs, but major evisceration is carried out by the lysosome. Lysosomes were discovered in 1949 by Christian de Duve, for which he won the 1974 Nobel Prize in Physiology or Medicine. 13 Lysosomes are highly acidic (pH 4.8) vesicular structures that serve as the cell’s primary waste disposal system. Sometimes called “suicide-bags,” these globules fuse with and release hydrolytic enzymes into the autophagic vacuoles which engulf malfunctioning cellular structures.
Certain pathogenic molecules build up at a rate that indicates they are impervious or highly resistant to catabolism. Some enzymes are rate-limiting – for example, when deconstructing a car, one needs many different tools in sequence. The lack of any one of them can halt the process. This fact is a double-edged sword, however: although numerous enzymes are involved in any given metabolic pathway, it may be the case that few will be required to transform the problem substance into a compound that the mammalian metabolism can already handle. It is a matter of tipping the scale in the patient’s favor, not guiding the entire reaction from start to finish.
Lysosomes are fallible. Certain molecules, like lipofuscin or “age pigment” builds up within clogged lysosomes called residual bodies. This process progresses at a set rate during “normal” aging – some medical conditions arise due to a faster rate of aggregation. These aberrant lysosomes can be supplemented with catabolic enzymes.
Lysosomal Storage Diseases
Lysosomal malfunction can lead to lysosomal storage diseases (LSDs), which are a group of genetically recessive disorders resulting from deficiencies of acid hydrolases. They have a prevalence of 1 per 7700 live births and constitute a significant burden on society. These include disorders such as Tay-Sachs, Gaucher’s, Pompe’s, Krabbe’s and Fabry’s diseases, which are treated with intravenous enzyme replacement therapy (ERT). 14 In the future, these conditions may be treated with gene therapy. LSDs are relevant to medical bioremediation for other conditions as a proof-of-concept for enzyme supplementation.
Medical Applications
Lysosomal catabolic insufficiency is involved, to varying degrees, in three major degenerative diseases. Atherosclerosis, macular degeneration and various neurodegenerative conditions (including Alzheimer's dementia) all feature the buildup of intra- and inter-cellular aggregates. These pathogenic aggregates can be degraded when the appropriate exogenous enzymes are supplemented.
Pathophysiology of Common Degenerative Diseases
Atherosclerosis
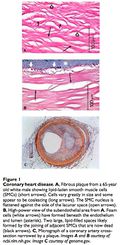
Atherosclerosis is the condition of arterial hardening due to plaque buildup. This process may result in the formation of a thrombus or clot that occludes blood flow (stroke or heart attack). Beginning in childhood, 15 atherosclerotic plaques slowly form on the endothelium of blood vessels. These plaques are composed of cholesterol, triglycerides and foam cells. Cholesterol and triglycerides are bound to and deposited by low-density lipoproteins (LDL) and are carried away by high-density lipoproteins (HDL). The “unstable” plaques most prone to rupture are inflamed and composed of macrophages and foam cells rather than the “stable” plaques composed of more extracellular matrix and smooth muscle cells. The LDL molecules in the atheroma become toxic after being oxidized by free radicals, triggering immune cells to migrate to the site. Macrophages and T-lymphocytes are unable to catabolize the LDL and become lipid-loaded foam cells. The lysosomes of these foam cells are precisely what fill with lipids. 16, 17 Lysosomes require a low pH for enzymatic function; this is established by vacuolar ATPases (proton pump). The overabundance of free cholesterol within the foam cells may actually inhibit the proton pumps, generating a positive feedback loop. 18 Lipase enzymes are responsible for triglyceride degradation and could be applied in enzyme replacement therapy. 19
Age-Related Macular Degeneration
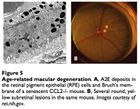
Macular degeneration is an age-related condition characterized by impaired sight in the center of vision (macula) due to damage to the retina. There are two types of AMD, the first being rarer and also more tractable: wet and dry. The first involves neovascularization that occludes vision and can be treated with anti-VEGF antibodies (VEGF being an angiogenesis factor). There are no commonly accepted treatments for dry AMD, perhaps other than antioxidant supplementation to prevent further damage to retinal pigment epithelium cells.
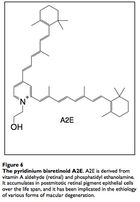
The input signal to vision is light in the form of photons. Photons are absorbed by the compound retinal, which transmits the signal into electrical impulses carried by the optic nerve and then to the visual cortex. The retinal itself stereoisomerises from 11-cis-retinal to all-trans-retinal, then to all-trans-retinol to 11-cis-retinol and back to 11-cis-retinal. Occassionally, all-trans-retinal reacts with the membrane phospholipid phosphatidylethanolamine to form a compound called A2E that builds up in the photoreceptor cells and retinal pigmented epithelial cells (which conduct lysosomal recycling). A2E cannot be catabolized by any enzyme and builds up in the RPE cells, constituting 20% of cellular mass by old age,20 leading to vision issues. Engineering microbes to produce an enzyme capable of catabolizing A2E may attenuate the costly effects of macular degeneration, and some peroxidases have proven capable of A2E degradation. 21
Neurodegeneration
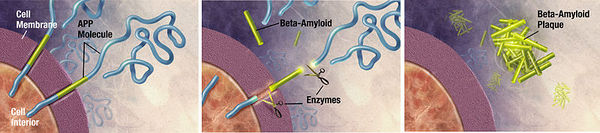
Unwanted molecules also build up in diseases of the nervous system. Alzheimer’s dementia, an emerging epidemic, is characterized by the presence of two culprits: a 40-43 amino acid extracellular amyloid-β peptide and the 352-441 amino acid hyperphosphorylated, microtubule-associated tau proteins which form neurofibrillary tangles.22 α-synuclein is another peptide putatively involved with both dementia and Parkinson’s disease. Huntingtin is a peptide aggregate involved with Huntington’s disease. Various neural pathologies are caused or exacerbated by the buildup of particular peptide molecules into aggrosomes. 23 It has been demonstrated that aggrosomes can be degraded by glial cells called Schwann cells. 24 Furthermore, lysosomal turnover can be pharmacologically stimulated to reduce aggregate accumulation and ameliorate neurological deficits in mice.25 Proteolytic enzyme supplementation could again be used to treat neurological conditions caused by unwanted peptide aggregation.
Microbial Engineering Advancements
Microbes have proven capable of degrading recalcitrant cellular aggregates in preliminary studies. Organisms like Nocardia nova, Chromobacterium sp. DS1, and Mycoplasma hyorhinis have proven capable of degrading such pathogenic molecules as 7-ketocholesterol and amyloid beta.
Microbial Degradation of 7-Ketocholesterol
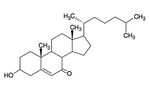
7-ketocholesterol (7KC) is an oxidized derivative of cholesterol, a plasma membrane stabilizer and hormone precursor. 7KC is suspected of involvement in both atherosclerosis,26 macular degeneration 27and Alzheimer’s 28 as a pro-oxidant and pro-inflammatory agent. 29 7KC is the most commonly consumed dietary oxysterol and has been shown to accumulate in human plasma. 30 Mammalian enzymes are capable of degrading 7KC, but they’re not found in the lysosome.
Bacteria do not have cholesterol, they have structurally similar “surrogate” compounds called hopanoids, which confer membrane stability in a similar fashion. 31 Despite this absence, Mathieu et al. (2008) demonstrated that several microbial species are capable of degrading cholesterol as a sole carbon source, including γ-Proteobacterium Y-134, Sphingomonas sp. JEM-1, Nocardia nova, Rhodococcus sp. RHA1 and Pseduomonas aeruginosa. 32, 33 Nocardia nova was most effective, degrading nearly 100% of the 7KC substrate in 25 days (30°C, pH 7). The runners up, γ – Proteobacterium and Sphingomonas were only able to catabolize about 80% at 25 days. This discrepancy may be due to Nocardia’s ability to produce biosurfactants 34 that dissolve the hydrophobic 7KC and increase the surface area available for enzymatic binding.
These microbes possess oxygenase enzymes that bind molecular oxygen with hydrogen and attach the hydroxyl groups onto highly reduced aromatics. As these new hydroxyl groups are attached, the molecule becomes susceptible to dehydrogenase and hydroxylase attacks. 35 This may tip the scale and enable mammalian enzymes to act on the sterol substrate.
7KC resistance in fibroblasts transfected with a lysosomally targeted Chromobacterium oxidase
In oxidized LDL-loaded macrophage foam cells, which are a primary component of atherosclerotic plaques, approximately 54% of the free 7KC is inside the lysosome. 36
High levels of 7KC cause lysosomal membrane permeabilization (LMP), leading to leakage of protease enzymes (like cathepsins), apoptosis and necrosis. 37
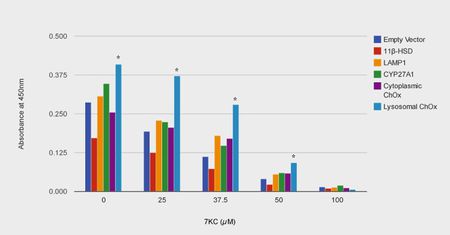
Mathieu et al. (2012) discovered that, unlike other enzymes capable of catabolizing 7KC, a novel cholesterol oxidase from Chromobacterium sp. DS1 was able to withstand the low pH, proteases and lack of cofactors that characterize the lysosome. 38
The authors localized DS1 ChOx to the lysosome by fusing Lysosomal Associated Membrane Protein 1 (LAMP1) to both the C terminus and N terminus of the cholesterol oxidase peptide.
At all but the highest interval of 7KC exposure (0, 25, 37.5, 50 µM but not 100 µM) the ChOx-LAMP1 construct afforded significant protection (maximum of 160% increase in cell viability at 37.5µM compared to control). Cytoplasmically targeted ChOx elicited a statistically insignificant level of protection. The authors conclude that this study “strongly suggests that reducing lysosomal oxysterol concentrations is promising for mitigating oxysterol toxicity.”
A2E is a major constituent of pigment lipofuscin, which builds in retinal epithelium cells with age. Dr. John Schloendorn and the team of Dr. Janet Sparrow collaborated to show that certain enzymes were capable of catabolizing the particularly sturdy A2E. These include: Crocus sativus zeaxanthin cleavage dioxygenase, cyanobacterial carotenoid cleavage dioxygenase NSC1, horseradish peroxidase (HRP) and several peroxidases. The researchers further showed that HRP could be targeted to human lysosomes in culture and there could non-toxically degrade A2E. 56, 57

Amyloid Beta Degradation by Mycoplasma hyorhinis
Amyloid beta is an extracellular aggregate protein putatively implicated in Alzheimer’s dementia. Zhao, H. et al. (2008) found that Mycoplasma hyorhinis is capable of degrading A-β. It is estimated that 5% to 35% of cell cultures in current use are infected with various mycoplasma species.39 Mycoplasmas are the smallest and simplest known self-replicating bacteria and the consequences of infection range from no apparent effect to apoptosis induction. The researchers observed no accumulation of A-β in mycoplasma-positive cells transfected with A-β precursor protein, whereas eradication of M. hyorhinis using a quinolone-based antibiotics restored extracellular A-β accumulation. 40 This experiment serves as another proof-of-concept that microbial ingenuity can be leveraged to degrade recalcitrant cellular aggregates like A-β.
Microbial Enzyme Screening
Leveraging the microbial infallibility hypothesis to find candidate genes and hydrolytic enzymes is the truly novel concept in medical bioremediation and microbial ecology is the key discipline. Since the 1970s, bioremediation has become more sophisticated, largely due to the realization that microbes are syntrophic, digesting compounds as a chain of hydrolytic, oxidative and reductive reactions across multiple species. These organisms also show redundancy, in that different species carry out the same steps using similar enzymes.
To harvest the critical xenoenzymes, the microbe must be identified and isolated. Given that less than 1% of microbial species are known and culturable, we must use techniques that allow both accuracy and speed. These techniques include molecular fingerprinting and DNA microarray analysis. Typically, 16S SSU rRNA is used to discern the phylogentic identity of a sample microbe. 16S rRNA is used because it is conserved across species and serves as an accurate molecular clock. 41
A phenol-chloroform extraction may be used to isolate nucleic acid. Then, newly identified microbes demonstrating the desired catabolic ability would be sequenced. Loss-of-function knockout methods (bioinformatics or random transposon-mediated mutagenesis) can illuminate DNA sequences that code for the target catabolic enzyme(s). Then, it may be possible to constitutively overexpress the enzyme-coding gene and recover the viable product.
Drug Delivery Methods and Obstacles
Drug Delivery Methods
Catabolic enzymes can only be effective when delivered to the target substrate. Fortunately, methods developed for the treatment of LSDs can be borrowed for the medical bioremediation of diseases not strictly due to lysosomal malfunction. In two common LSDs, the target cells are easily accessible to injected enzymes (in Gaucher’s the target is the macrophage and in Fabry’s it is the vascular endothelium). In order for specific cells to take up enzymes in circulation, they are encapsulated in a carrier that is glycosylated (conjugated with a polysaccharide) to be recognizable to lectin receptors in the target cell type. 42 Over 4,000 patients have been treated with ERT and the are now capable of living normal lives.43
The downside to ERT is that large quantities of expensive enzyme must be frequently administered because lysosomal enzymes themselves are rapidly degraded. Furthermore, the ever-stubborn blood-brain barrier adds a layer of difficulty when seeking to apply ERT to the brain. A conceptually obvious but still technically difficult solution would be to engineer the in situ synthesis of exogenous lysosomal enzymes via gene therapy or ex vivo stem cell transfection and subsequent implantation. 44
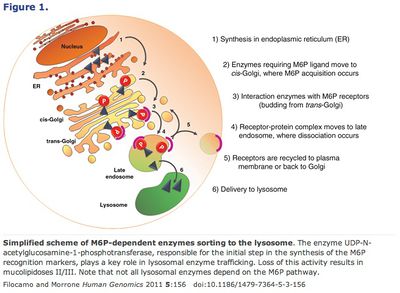
Alternatively, conjugating mannose-6-phosphate to the supplemental enzymes takes advantage of the lysosome’s own transmission pathway from the Golgi apparatus. 45 Finally, another method, chaperone-mediated autophagy (CMA) bypasses the Golgi entirely. This pathway relies on chaperone proteins (produced in the endoplasmic reticulum) to recognize specific amino acid motifs, bind them and then be translocated together by the lysosome receptor hsc70 (or HSPA8). 46
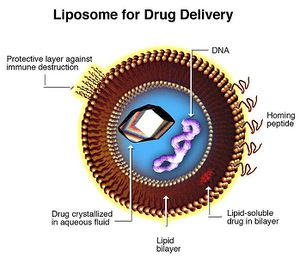
Liposomes hold great promise for xenoenzyme delivery. 47 Liposomes are artificial vesicles ranging from 200 to 20 nanometers. They can transport drugs across phospholipid membranes regardless of lipid solubility of the compound itself. Liposome membranes can be conjugated with various molecules to control systemic distribution. Conjugates include hydrophilic cloaking agents like polyethylene glycol; antibodies and homing peptides; stimulus-sensitive release triggering lipids and peptides (pH, temperature, ligand specific); imaging particles, fluorophores; gene therapy products and transfection vectors (DNA, RNA, siRNA, transposons, et cetera).
Side Effects and Obstacles
Inactivity and Toxicity
If the purified xenoenzymes are ineffective or toxic, the simple solution is to just keep looking for others that are less so. However, more directed methods may expedite the process. If the problem is due to the disparity between microbial and human intralysosomal pH, this is justification to investigate fungal hydrolases more assiduously, because fungi and humans have relatively similar lysosomal transport localization mechanisms and pH function ranges. 48
Toxicity will be defined as degradation of vital extralysosomal structures and/or physically impeding their movement. If this occurs, it is possible to synthesize the enzyme as a non-functional proenzyme that must be cleaved within the lysosome in order to function (this is how most mammalian enzymes already work).49
Immune Rejection
Once again, the trailblazing research in enzyme replacement therapy (ERT) sheds light on potential challenges to medical bioremediation. ERT patients are actually able to tolerate large doses of injected enzyme without eliciting an immune response (even with immunosuppressants). Moreover, reactivity tends to decline in those with sensitivities. 50 Because LSD patients are congenitally deficient, the supplemental enzyme is as equivalently “non-self” as would be microbial enzymes.14 The low level of immunogenicity is thought to be due to the fact that lysosomal enzymes are taken up from the cytosol and are not exposed to the immunoproteasome, and therefore digested fragments are not loaded on HLA complexes for cell surface presentation. Unfortunately, the circulating antibodies of the humoral immune response may still instigate an immune response. If a patient’s immune response were prohibitively serious, immunosuppressants may be justified.
Blood-Brain Barrier
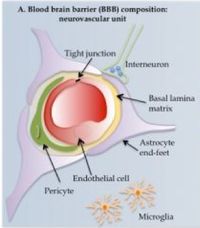
The blood brain barrier (BBB) is a physical filtration system that prevents potential toxins and pathogens from entering the central nervous system. Elsewhere in the body, large, polar molecules escape blood vessels through fenestrations or periodic gaps between endothelial cells. These are absent in the CNS (tight junctions). Furthermore, specialized glial cells called astrocytes have long appendages, end feet or processes that constitute another barrier layer. This BBB is an ever-present obstacle in neurology. Various methods of circumvention have been developed, however.
The prevailing method involves injecting high molarity mannitol to exert osmotic pressure on the endothelial cells. The high solute concentration (hypertonic) in the blood causes water to diffuse out of the endothelial cells, distorting them (flaccid) enough to open up the tight junctions and allow molecules to pass in an approximately five minute window.52 This method is indispensable for smallecule administration but does not work as well for large peptide transmission.
Transferrin receptor-mediated uptake is another viable method. Transferrin is an iron-binding peptide that regulates the level of free serum iron. The enzyme binds iron in the mucosa, depriving potential pathogens of iron and reducing survival in a process called iron withholding. The receptor is present in CNS endothelial cells. Conjugating the drug or peptide in question to transferrin allows for active transport into the nervous system.53, 54 Finally, another emerging method involves using liposomes to get across the BBB and transfect neurons and glia with the target gene. 55
Conclusion
Medical bioremediation research is ongoing. The goal is to safely degrade pathogenic, recalcitrant aggregates and ameliorate some of the most common and debilitating diseases. The field is still in its infancy, but great strides have been made as researchers from various, disparate disciplines like environmental microbiology and mammalian cellular biology have come together. Medical bioremediation is a disruptive technology and many institutional, theoretical and technical challenges remain. Further research directions include: Isolation of competent microbial strains and starvation selection, mutagenesis and enzyme identification, lysosomally-targeted transgene development, cell and in vivo toxicity assaying, and human trials.
References
1. Vidali, M. (2001) Bioremediation: An Overview. Pure Appl. Chem. 73 (7): 1163–1172.
2. Brim H. et al. (2000). Engineering Deinococcus radiodurans for metal remediation in radioactive mixed waste environments. Nat. Biotechnol. 18 (1): 85–90.
3. Demain, A.L. (2009) Antibiotics: natural products essential for human health. Med. Res. Rev. 29: 821–841
4. Leader, B. et al. (2008) Protein therapeutics: a summary and pharmacological classification. Nat. Rev. Drug Discov. 7(1): 21–39
5. Gale, E.F. (1952). The Chemical Activities of Bacteria. New York, Academic Press.
6. de Grey, A. et al. (2005) Medical bioremediation: Prospects for the application of microbial catabolic diversity to aging and several major age-related diseases. Ageing Res Rev. 4(3):315-38.
7. de Grey, A. (2005). A strategy for postponing aging indefinitely. Stud Health Technol Inform. 118: 209–19.
8. de Grey, A. et al. (2004). Total deletion of in vivo telomere elongation capacity: an ambitious but possibly ultimate cure for all age-related human cancers. Ann NY Acad Sci. 1019: 147–70.
9. de Grey, A. (2006). Foreseeable pharmaceutical repair of age-related extracellular damage. Curr Drug Targets. 7 (11): 1469–77.
10. de Grey, A. (2004). Mitochondrial Mutations in Mammalian Aging: An Over-Hasty About-Turn? Rejuvenation Res. 7 (3): 171–4.
11. de Grey, A. (2002). Bioremediation meets biomedicine: therapeutic translation of microbial catabolism to the lysosome. Trends Biotechnol. 20: 452–455.
12. Furber, J.D. (2006). Extracellular glycation crosslinks: Prospects for removal. Rejuvenation Res. 9 (2): 274–278.
13. Bowers, W.E. (1998) Christian de Duve and the discovery of lysosomes and peroxisomes. Trends Cell Biol. 8: 330–333
14. Schiffmann, R. et al. (2001). Enzyme replacement therapy in Fabry disease: a randomized controlled trial. JAMA 285: 2743–2749.
15. Strong, J.P. et al. (1958). The natural history of the early aortic lesions in New Orleans, Guatemala, and Costa Rico. Am. J. Pathol. 34: 731–744.
16. Jerome W.G. (2006) Advanced atherosclerotic foam cell formation has features of an acquired lysosomal storage disorder. Rejuvenation Res. 9(2):245-55.
17. Jerome, W.G. and Yancey, P.G. (2003). The role of microscopy in understanding atherosclerotic lysosomal lipid metabolism. Microsc. Microanal. 9: 54–67.
18. D’Souza, M.P. et al. (1987). Reconstitution of the lysosomal proton pump. Proc Natl Acad Sci. 84: 6980–6984.
19. Jerome WG et al. (2008) Lysosomal cholesterol accumulation inhibits subsequent hydrolysis of lipoprotein cholesteryl ester. Microsc Microanal. 14(2):138-49.
20. Feeney-Burns, L. et al. (1984). Aging human RPE: morphometric analysis of macular, equatorial, and peripheral cells. Invest. Ophthalmol. Vis. Sci. 25: 195–200.
21. Schloendorn J. et al. (2009) Medical Bioremediation: A Concept Moving Toward Reality. Rejuvenation Res. 12(6):411-419.
22. Mattson, M.P. (2004). Pathways towards and away from Alzheimer’s disease. Nature. 430: 631–639.
23. Wood, J. et al. (2003). Protein aggregation in motor neurone disorders. Neuropathol. Appl. Neurobiol. 29, 529–545.
24. Fortun, J. et al. (2003). Emerging role for autophagy in the removal of aggresomes in Schwann cells. J. Neurosci. 23, 10672–10680.
25. Ravikumar, B. et al. (2004). Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 36: 585–595.
26. Hughes H. et al. (1994) Cytotoxicity of oxidized LDL to porcine aortic smooth muscle cells is associated with the oxysterols 7-ketocholesterol and 7-hydroxycholesterol. Arterioscler. Thromb. 14: 1177–1185
27. Rodriguez I.R. and Larrayoz I.M. (2010). Cholesterol oxidation in the retina: Implications of 7KCh formation in chronic inflammation and age-related macular degeneration. J. Lipid. Res. 51 (10): 2847–2862.
28. Nelson T.J. and Alkon D.L. (2005) Oxidation of cholesterol by amyloid precursor protein and beta-amyloid peptide. J Biol Chem 280:7377–7387
29. Dushkin M. et al. (1998) Effects of oxysterols upon macrophage and lymphocyte functions in vitro. Prostaglandins. Lipid Mediat. 55: 219–236
30. Linseisen J and Wolfram G (1998) Absorption of cholesterol oxidation products from ordinary foodstuff in humans. Ann. Nutr. Metab. 42:221–230
31. Sáenz J.P. et al. (2012) Functional convergence of hopanoids and sterols in membrane ordering. Proc. Natl. Acad. Sci. 109(35):14236-40.
32. Mathieu J. et al. (2008) Microbial degradation of 7-ketocholesterol. Biodegradation. 19:807–813
33. Van der Geize R. et al. (2007) A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc. Natl. Acad. Sci. 104:1947–1952
34. Margaritis A. et al. (1979) Biosurfactant production by Nocardia erythropolis. Dev. Ind. Microbiol. 20:623–630
35. Sligar SG et al. (2005). Thirty years of microbial P450 monooxygenase research: peroxo-heme intermediates--the central bus station in heme oxygenase catalysis. Biochem. Biophys. Res. Commun. 338 (1): 346–54
36. Brown AJ et al. (2000). Cholesterol and oxysterol metabolism and subcellular distribution in macrophage foam cells. Accumulation of oxidized esters in lysosomes. J. Lipid. Res. 41(2):226–237.
37. Boya P and Kroemer G. (2008) Lysosomal membrane permeabilization in cell death. Oncogene. 50: 6434-51
38. Mathieu JM et al. (2012) Increased Resistance to Oxysterol Cytotoxicity in Fibroblasts Transfected With a Lysosomally Targeted Chromobacterium Oxidase. Biotechnol Bioeng. 109(9):2409-15.
39. Young L. et al. (2010) Detection of Mycoplasma in cell cultures. Nature Protocols. 5(5); 929–934.
40. Zhao, H. et al. (2008) Amyloid-beta peptide degradation in cell cultures by mycoplasma contaminants. BMC Res. Notes. 1(1);38
41. Woese, C.R. (1987). Bacterial evolution. Microbiology Reviews. 51; 221–271.
42. Das, P.K. et al. (1985). Lectin-specific targeting of b-glucocerebrosidase to different liver cells via glycosylated liposomes. Biochem. Med. 33: 124–131
43. Brady, R.O. (2003). Gaucher and Fabry diseases: from understanding pathophysiology to rational therapies. Acta Paediatr. Suppl. 92: 19–24.
44. Lai, Z. and Brady, R.O. (2002). Gene transfer into the central nervous system in vivo using a recombinant lentivirus vector. J. Neurosci. Res. 67: 363–371.
45. Ghosh, P. et al. (2003). Mannose 6-phosphate receptors: new twists in the tale. Nat. Rev. Mol. Cell Biol. 4: 202–212
46. Massey, A.C. (2004). Pathophysiology of chaperone-mediated autophagy. Int. J.Biochem. Cell Biol. 36: 2420–2434.
47. Torchilin V.P. (2005) Recent advances with liposomes as pharmaceutical carriers. Nat. Rev. Drug Deliv. 4:145-160.
48. Levine, B. and Klionsky, D.J., (2004). Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6, 463–477.
49. Wittlin, S. et al. (1999). Mechanisms and kinetics of procathepsin D activation. Eur. J. Biochem. 265, 384–393.
50. Brooks, D. et al. (2003). Significance of immune response to enzyme-replacement therapy for patients with a lysosomal storage disorder. Trends Mol. Med. 9, 450–453.
51. Ikeda, M. et al. (2002) Synergistic Effect of Cold Mannitol and Na+/Ca2+ Exchange Blocker on Blood-Brain Barrier Opening. Biochem. Biophys. Res. Commun. 291, 669–674.
52. Bickel, U. et al. (2001). Delivery of peptides and proteins through the blood–brain barrier. Adv. Drug Deliv. Rev. 46: 247–279.
53. Qian ZM et al. (2002). Targeted drug delivery via the transferrin receptor-mediated endocytosis pathway. Pharmacol. Rev. 54(4): 561-87.
54. Schnyder A. and Huwyler J. (2005) Drug Transport to Brain with Targeted Liposomes. NeuroRx. 2(1): 99–107.
55. Filocamo M. and Morrone A. (2011) Lysosomal storage disorders: Molecular basis and laboratory testing. Hum. Genom. 5(3): 156–169.
56. Schloendorn, J. (2009) Progress Towards Medical Bioremediation by Enzymatic Transformation of 7-ketocholesterol and the pyridinium bisretinoid A2E. PhD Dissertation, Arizona State University. 3392127.
57. Sparrow et al. (2011) Enzymatic degradation of A2E, a retinal pigment epithelial lipofuscin bisretinoid. J Am Chem Soc. 133(4):849-57
Edited by Sebastian Aguiar, a student of Nora Sullivan in BIOL187S (Microbial Life) in The Keck Science Department of the Claremont Colleges Spring 2013.